BASIRA Lab
Brain graph synthesis marked a new era for predicting a target brain graph from a source one without incurring the high acquisition cost and processing time of neuroimaging data. However, existing multi-modal graph synthesis frameworks have several limitations. First, they mainly focus on generating graphs from the same domain (intra-modality), overlooking the rich multimodal representations of brain connectivity (inter-modality). Second, they can only handle isomorphic graph generation tasks, limiting their generalizability to synthesizing target graphs with a different node size and topological structure from those of the source one. More importantly, both target and source domains might have different distributions, which causes a domain fracture between them (i.e., distribution misalignment). To address such challenges, we propose an inter-modality aligner of non-isomorphic graphs (IMANGraphNet) framework to infer a target graph modality based on a given modality. Our three core contributions lie in (i) predicting a target graph (e.g., functional) from a source graph (e.g., morphological) based on a novel graph generative adversarial network (gGAN); (ii) using non-isomorphic graphs for both source and target domains with a different number of nodes, edges and structure; and (iii) enforcing the predicted target distribution to match that of the ground truth graphs using a graph autoencoder to relax the designed loss oprimization. To handle the unstable behavior of gGAN, we design a new Ground Truth-Preserving (GT-P) loss function to guide the generator in learning the topological structure of ground truth brain graphs. Our comprehensive experiments on predicting functional from morphological graphs demonstrate the outperformance of IMANGraphNet in comparison with its variants. This can be further leveraged for integrative and holistic brain mapping in health and disease.
Metadata-Driven Federated Learning of Connectional Brain Templates in Non-IID Multi-Domain Scenarios
Metadata-Driven Federated Learning of Connectional Brain Templates in Non-IID Multi-Domain Scenarios
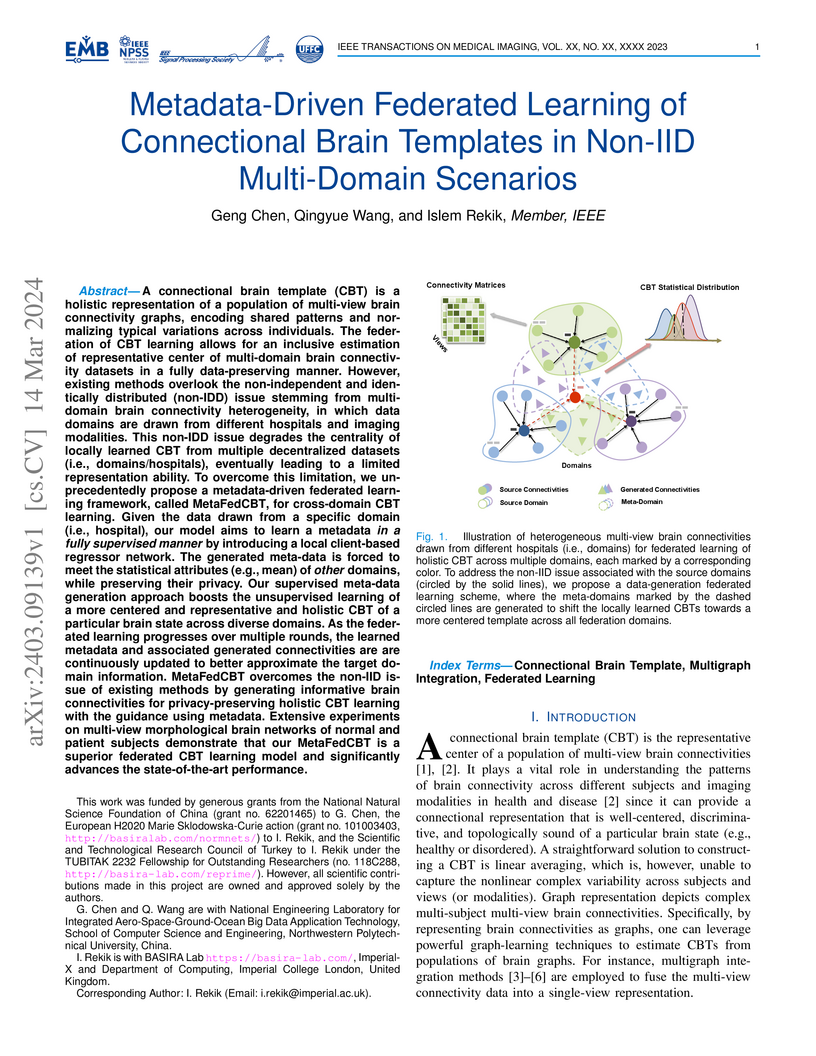
A connectional brain template (CBT) is a holistic representation of a
population of multi-view brain connectivity graphs, encoding shared patterns
and normalizing typical variations across individuals. The federation of CBT
learning allows for an inclusive estimation of the representative center of
multi-domain brain connectivity datasets in a fully data-preserving manner.
However, existing methods overlook the non-independent and identically
distributed (non-IDD) issue stemming from multidomain brain connectivity
heterogeneity, in which data domains are drawn from different hospitals and
imaging modalities. To overcome this limitation, we unprecedentedly propose a
metadata-driven federated learning framework, called MetaFedCBT, for
cross-domain CBT learning. Given the data drawn from a specific domain (i.e.,
hospital), our model aims to learn metadata in a fully supervised manner by
introducing a local client-based regressor network. The generated meta-data is
forced to meet the statistical attributes (e.g., mean) of other domains, while
preserving their privacy. Our supervised meta-data generation approach boosts
the unsupervised learning of a more centered, representative, and holistic CBT
of a particular brain state across diverse domains. As the federated learning
progresses over multiple rounds, the learned metadata and associated generated
connectivities are continuously updated to better approximate the target domain
information. MetaFedCBT overcomes the non-IID issue of existing methods by
generating informative brain connectivities for privacy-preserving holistic CBT
learning with guidance using metadata. Extensive experiments on multi-view
morphological brain networks of normal and patient subjects demonstrate that
our MetaFedCBT is a superior federated CBT learning model and significantly
advances the state-of-the-art performance.
Deep learning models often struggle to maintain generalizability in medical imaging, particularly under domain-fracture scenarios where distribution shifts arise from varying imaging techniques, acquisition protocols, patient populations, demographics, and equipment. In practice, each hospital may need to train distinct models - differing in learning task, width, and depth - to match local data. For example, one hospital may use Euclidean architectures such as MLPs and CNNs for tabular or grid-like image data, while another may require non-Euclidean architectures such as graph neural networks (GNNs) for irregular data like brain connectomes. How to train such heterogeneous models coherently across datasets, while enhancing each model's generalizability, remains an open problem. We propose unified learning, a new paradigm that encodes each model into a graph representation, enabling unification in a shared graph learning space. A GNN then guides optimization of these unified models. By decoupling parameters of individual models and controlling them through a unified GNN (uGNN), our method supports parameter sharing and knowledge transfer across varying architectures (MLPs, CNNs, GNNs) and distributions, improving generalizability. Evaluations on MorphoMNIST and two MedMNIST benchmarks - PneumoniaMNIST and BreastMNIST - show that unified learning boosts performance when models are trained on unique distributions and tested on mixed ones, demonstrating strong robustness to unseen data with large distribution shifts. Code and benchmarks: this https URL
There are no more papers matching your filters at the moment.