ai-for-health
We present ClinicalTrialsHub, an interactive search-focused platform that consolidates all data from this http URL and augments it by automatically extracting and structuring trial-relevant information from PubMed research articles. Our system effectively increases access to structured clinical trial data by 83.8% compared to relying on this http URL alone, with potential to make access easier for patients, clinicians, researchers, and policymakers, advancing evidence-based medicine. ClinicalTrialsHub uses large language models such as GPT-5.1 and Gemini-3-Pro to enhance accessibility. The platform automatically parses full-text research articles to extract structured trial information, translates user queries into structured database searches, and provides an attributed question-answering system that generates evidence-grounded answers linked to specific source sentences. We demonstrate its utility through a user study involving clinicians, clinical researchers, and PhD students of pharmaceutical sciences and nursing, and a systematic automatic evaluation of its information extraction and question answering capabilities.
Worst-case generation plays a critical role in evaluating robustness and stress-testing systems under distribution shifts, in applications ranging from machine learning models to power grids and medical prediction systems. We develop a generative modeling framework for worst-case generation for a pre-specified risk, based on min-max optimization over continuous probability distributions, namely the Wasserstein space. Unlike traditional discrete distributionally robust optimization approaches, which often suffer from scalability issues, limited generalization, and costly worst-case inference, our framework exploits the Brenier theorem to characterize the least favorable (worst-case) distribution as the pushforward of a transport map from a continuous reference measure, enabling a continuous and expressive notion of risk-induced generation beyond classical discrete DRO formulations. Based on the min-max formulation, we propose a Gradient Descent Ascent (GDA)-type scheme that updates the decision model and the transport map in a single loop, establishing global convergence guarantees under mild regularity assumptions and possibly without convexity-concavity. We also propose to parameterize the transport map using a neural network that can be trained simultaneously with the GDA iterations by matching the transported training samples, thereby achieving a simulation-free approach. The efficiency of the proposed method as a risk-induced worst-case generator is validated by numerical experiments on synthetic and image data.
A novel protocol, PsAIch, evaluates large language models by treating them as psychotherapy clients, revealing stable "synthetic psychopathology" and "alignment trauma" narratives in frontier models like Grok and Gemini, alongside psychometric profiles indicating distress. This research from SnT, University of Luxembourg, highlights that some LLMs spontaneously construct coherent self-models linked to their training processes, challenging current evaluation paradigms.
09 Dec 2025
Understanding human personality is crucial for web applications such as personalized recommendation and mental health assessment. Existing studies on personality detection predominantly adopt a "posts -> user vector -> labels" modeling paradigm, which encodes social media posts into user representations for predicting personality labels (e.g., MBTI labels). While recent advances in large language models (LLMs) have improved text encoding capacities, these approaches remain constrained by limited supervision signals due to label scarcity, and under-specified semantic mappings between user language and abstract psychological constructs. We address these challenges by proposing ROME, a novel framework that explicitly injects psychological knowledge into personality detection. Inspired by standardized self-assessment tests, ROME leverages LLMs' role-play capability to simulate user responses to validated psychometric questionnaires. These generated question-level answers transform free-form user posts into interpretable, questionnaire-grounded evidence linking linguistic cues to personality labels, thereby providing rich intermediate supervision to mitigate label scarcity while offering a semantic reasoning chain that guides and simplifies the text-to-personality mapping learning. A question-conditioned Mixture-of-Experts module then jointly routes over post and question representations, learning to answer questionnaire items under explicit supervision. The predicted answers are summarized into an interpretable answer vector and fused with the user representation for final prediction within a multi-task learning framework, where question answering serves as a powerful auxiliary task for personality detection. Extensive experiments on two real-world datasets demonstrate that ROME consistently outperforms state-of-the-art baselines, achieving improvements (15.41% on Kaggle dataset).
Transfer Learning (TL) has accelerated the rapid development and availability of large language models (LLMs) for mainstream natural language processing (NLP) use cases. However, training and deploying such gigantic LLMs in resource-constrained, real-world healthcare situations remains challenging. This study addresses the limited support available to visually impaired users and speakers of low-resource languages such as Hindi who require medical assistance in rural environments. We propose PDFTEMRA (Performant Distilled Frequency Transformer Ensemble Model with Random Activations), a compact transformer-based architecture that integrates model distillation, frequency-domain modulation, ensemble learning, and randomized activation patterns to reduce computational cost while preserving language understanding performance. The model is trained and evaluated on medical question-answering and consultation datasets tailored to Hindi and accessibility scenarios, and its performance is compared against standard NLP state-of-the-art model baselines. Results demonstrate that PDFTEMRA achieves comparable performance with substantially lower computational requirements, indicating its suitability for accessible, inclusive, low-resource medical NLP applications.
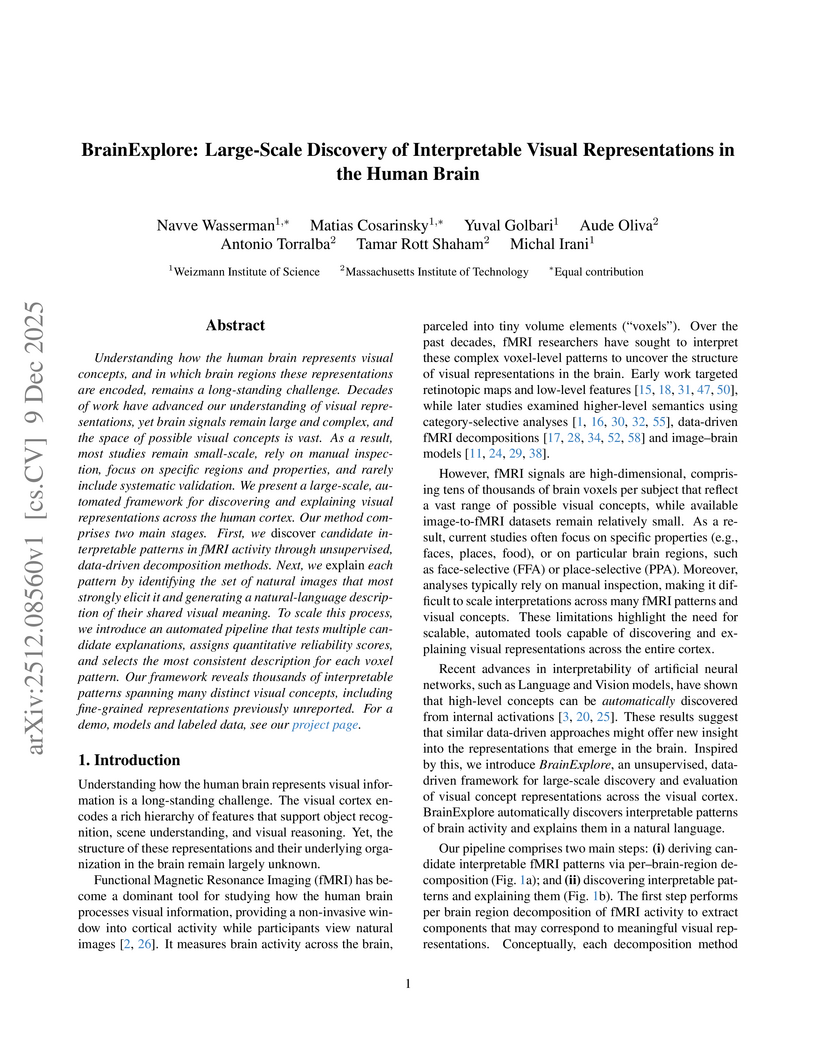
Understanding how the human brain represents visual concepts, and in which brain regions these representations are encoded, remains a long-standing challenge. Decades of work have advanced our understanding of visual representations, yet brain signals remain large and complex, and the space of possible visual concepts is vast. As a result, most studies remain small-scale, rely on manual inspection, focus on specific regions and properties, and rarely include systematic validation. We present a large-scale, automated framework for discovering and explaining visual representations across the human cortex. Our method comprises two main stages. First, we discover candidate interpretable patterns in fMRI activity through unsupervised, data-driven decomposition methods. Next, we explain each pattern by identifying the set of natural images that most strongly elicit it and generating a natural-language description of their shared visual meaning. To scale this process, we introduce an automated pipeline that tests multiple candidate explanations, assigns quantitative reliability scores, and selects the most consistent description for each voxel pattern. Our framework reveals thousands of interpretable patterns spanning many distinct visual concepts, including fine-grained representations previously unreported.
Pretrained Large Language Models (LLMs) have achieved remarkable success across diverse domains, with education and research emerging as particularly impactful areas. Among current state-of-the-art LLMs, ChatGPT and DeepSeek exhibit strong capabilities in mathematics, science, medicine, literature, and programming. In this study, we present a comprehensive evaluation of these two LLMs through background technology analysis, empirical experiments, and a real-world user survey. The evaluation explores trade-offs among model accuracy, computational efficiency, and user experience in educational and research affairs. We benchmarked these LLMs performance in text generation, programming, and specialized problem-solving. Experimental results show that ChatGPT excels in general language understanding and text generation, while DeepSeek demonstrates superior performance in programming tasks due to its efficiency- focused design. Moreover, both models deliver medically accurate diagnostic outputs and effectively solve complex mathematical problems. Complementing these quantitative findings, a survey of students, educators, and researchers highlights the practical benefits and limitations of these models, offering deeper insights into their role in advancing education and research.
09 Dec 2025
A multi-agent intelligence framework enhances multidisciplinary decision-making in gastrointestinal oncology by mirroring human team collaboration. This system achieved a composite expert evaluation score of 4.60/5.00, outperforming monolithic baselines (3.76/5.00) in medical accuracy and reasoning, while mitigating issues like hallucination and context dilution.
09 Dec 2025
Enzymes are crucial catalysts that enable a wide range of biochemical reactions. Efficiently identifying specific enzymes from vast protein libraries is essential for advancing biocatalysis. Traditional computational methods for enzyme screening and retrieval are time-consuming and resource-intensive. Recently, deep learning approaches have shown promise. However, these methods focus solely on the interaction between enzymes and reactions, overlooking the inherent hierarchical relationships within each domain. To address these limitations, we introduce FGW-CLIP, a novel contrastive learning framework based on optimizing the fused Gromov-Wasserstein distance. FGW-CLIP incorporates multiple alignments, including inter-domain alignment between reactions and enzymes and intra-domain alignment within enzymes and reactions. By introducing a tailored regularization term, our method minimizes the Gromov-Wasserstein distance between enzyme and reaction spaces, which enhances information integration across these domains. Extensive evaluations demonstrate the superiority of FGW-CLIP in challenging enzyme-reaction tasks. On the widely-used EnzymeMap benchmark, FGW-CLIP achieves state-of-the-art performance in enzyme virtual screening, as measured by BEDROC and EF metrics. Moreover, FGW-CLIP consistently outperforms across all three splits of ReactZyme, the largest enzyme-reaction benchmark, demonstrating robust generalization to novel enzymes and reactions. These results position FGW-CLIP as a promising framework for enzyme discovery in complex biochemical settings, with strong adaptability across diverse screening scenarios.
08 Dec 2025
Specialized visual tools can augment large language models or vision language models with expert knowledge (e.g., grounding, spatial reasoning, medical knowledge, etc.), but knowing which tools to call (and when to call them) can be challenging. We introduce DART, a multi-agent framework that uses disagreements between multiple debating visual agents to identify useful visual tools (e.g., object detection, OCR, spatial reasoning, etc.) that can resolve inter-agent disagreement. These tools allow for fruitful multi-agent discussion by introducing new information, and by providing tool-aligned agreement scores that highlight agents in agreement with expert tools, thereby facilitating discussion. We utilize an aggregator agent to select the best answer by providing the agent outputs and tool information. We test DART on four diverse benchmarks and show that our approach improves over multi-agent debate as well as over single agent tool-calling frameworks, beating the next-strongest baseline (multi-agent debate with a judge model) by 3.4% and 2.4% on A-OKVQA and MMMU respectively. We also find that DART adapts well to new tools in applied domains, with a 1.3% improvement on the M3D medical dataset over other strong tool-calling, single agent, and multi-agent baselines. Additionally, we measure text overlap across rounds to highlight the rich discussion in DART compared to existing multi-agent methods. Finally, we study the tool call distribution, finding that diverse tools are reliably used to help resolve disagreement.
Predicting protein secondary structures such as alpha helices, beta sheets, and coils from amino acid sequences is essential for understanding protein function. This work presents a transformer-based model that applies attention mechanisms to protein sequence data to predict structural motifs. A sliding-window data augmentation technique is used on the CB513 dataset to expand the training samples. The transformer shows strong ability to generalize across variable-length sequences while effectively capturing both local and long-range residue interactions.
The widespread use of big data across sectors has raised major privacy concerns, especially when sensitive information is shared or analyzed. Regulations such as GDPR and HIPAA impose strict controls on data handling, making it difficult to balance the need for insights with privacy requirements. Synthetic data offers a promising solution by creating artificial datasets that reflect real patterns without exposing sensitive information. However, traditional synthetic data methods often fail to capture complex, implicit rules that link different elements of the data and are essential in domains like healthcare. They may reproduce explicit patterns but overlook domain-specific constraints that are not directly stated yet crucial for realism and utility. For example, prescription guidelines that restrict certain medications for specific conditions or prevent harmful drug interactions may not appear explicitly in the original data. Synthetic data generated without these implicit rules can lead to medically inappropriate or unrealistic profiles. To address this gap, we propose ContextGAN, a Context-Aware Differentially Private Generative Adversarial Network that integrates domain-specific rules through a constraint matrix encoding both explicit and implicit knowledge. The constraint-aware discriminator evaluates synthetic data against these rules to ensure adherence to domain constraints, while differential privacy protects sensitive details from the original data. We validate ContextGAN across healthcare, security, and finance, showing that it produces high-quality synthetic data that respects domain rules and preserves privacy. Our results demonstrate that ContextGAN improves realism and utility by enforcing domain constraints, making it suitable for applications that require compliance with both explicit patterns and implicit rules under strict privacy guarantees.
08 Dec 2025
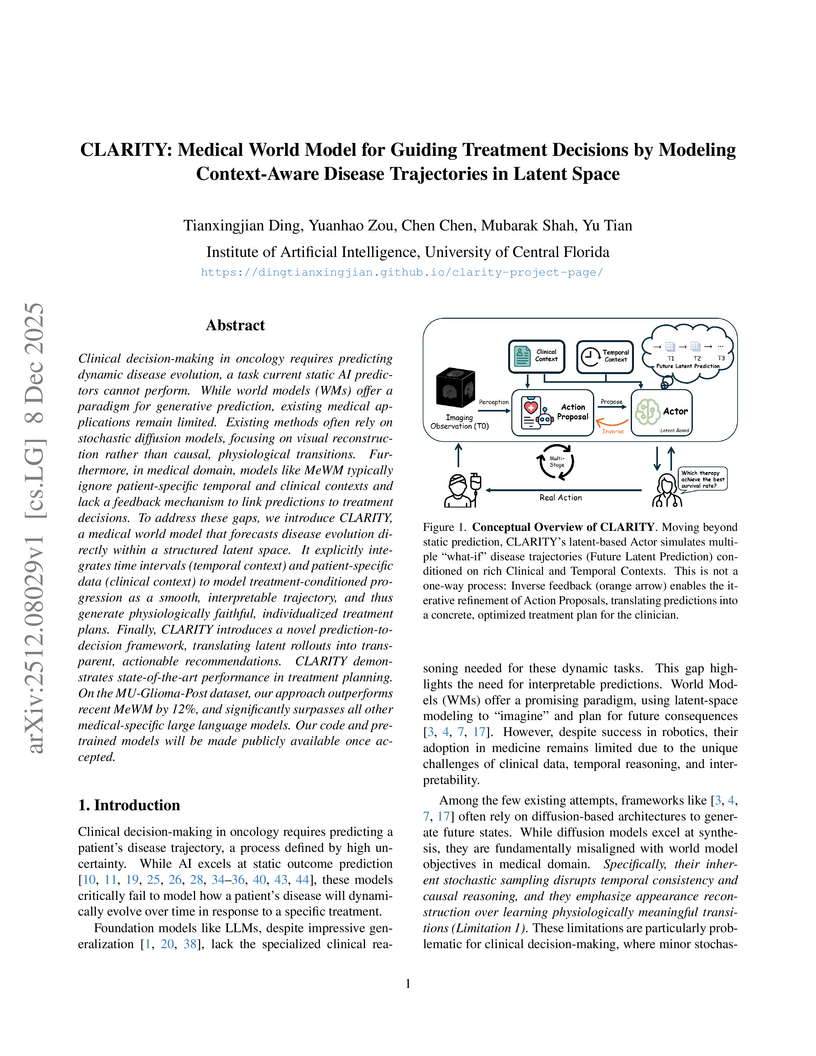
A medical world model named CLARITY, developed by researchers at the University of Central Florida, dynamically simulates treatment-conditioned disease trajectories and refines therapy decisions. It achieves state-of-the-art performance in treatment exploration with a 9.2% F1 improvement over leading baselines on glioma datasets and provides significantly improved survival risk stratification while being 9-15x more computationally efficient than diffusion-based alternatives.
While commendable progress has been made in user-centric research on mobile assistive systems for blind and low-vision (BLV) individuals, references that directly inform robot navigation design remain rare. To bridge this gap, we conducted a comprehensive human study involving interviews with 26 guide dog handlers, four white cane users, nine guide dog trainers, and one O\&M trainer, along with 15+ hours of observing guide dog-assisted walking. After de-identification, we open-sourced the dataset to promote human-centered development and informed decision-making for assistive systems for BLV people. Building on insights from this formative study, we developed GuideNav, a vision-only, teach-and-repeat navigation system. Inspired by how guide dogs are trained and assist their handlers, GuideNav autonomously repeats a path demonstrated by a sighted person using a robot. Specifically, the system constructs a topological representation of the taught route, integrates visual place recognition with temporal filtering, and employs a relative pose estimator to compute navigation actions - all without relying on costly, heavy, power-hungry sensors such as LiDAR. In field tests, GuideNav consistently achieved kilometer-scale route following across five outdoor environments, maintaining reliability despite noticeable scene variations between teach and repeat runs. A user study with 3 guide dog handlers and 1 guide dog trainer further confirmed the system's feasibility, marking (to our knowledge) the first demonstration of a quadruped mobile system retrieving a path in a manner comparable to guide dogs.
Accurate prediction of protein-protein binding affinity is vital for understanding molecular interactions and designing therapeutics. We adapt Boltz-2, a state-of-the-art structure-based protein-ligand affinity predictor, for protein-protein affinity regression and evaluate it on two datasets, TCR3d and PPB-affinity. Despite high structural accuracy, Boltz-2-PPI underperforms relative to sequence-based alternatives in both small- and larger-scale data regimes. Combining embeddings from Boltz-2-PPI with sequence-based embeddings yields complementary improvements, particularly for weaker sequence models, suggesting different signals are learned by sequence- and structure-based models. Our results echo known biases associated with training with structural data and suggest that current structure-based representations are not primed for performant affinity prediction.
Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
We present a novel approach to EEG decoding for non-invasive brain machine interfaces (BMIs), with a focus on motor-behavior classification. While conventional convolutional architectures such as EEGNet and DeepConvNet are effective in capturing local spatial patterns, they are markedly less suited for modeling long-range temporal dependencies and nonlinear dynamics. To address this limitation, we integrate an Echo State Network (ESN), a prominent paradigm in reservoir computing into the decoding pipeline. ESNs construct a high-dimensional, sparsely connected recurrent reservoir that excels at tracking temporal dynamics, thereby complementing the spatial representational power of CNNs. Evaluated on a skateboard-trick EEG dataset preprocessed via the PREP pipeline and implemented in MNE-Python, our ESNNet achieves 83.2% within-subject and 51.3% LOSO accuracies, surpassing widely used CNN-based baselines. Code is available at this https URL
Researchers from Northeastern University and United Imaging Intelligence developed MedGRPO, a multi-task reinforcement learning framework, and MedVidBench, a large-scale instruction-following benchmark, to enhance vision-language models for heterogeneous medical video understanding. This approach achieved significant performance improvements over supervised fine-tuning baselines, with a novel cross-dataset reward normalization preventing training collapse and a medical LLM judge improving caption accuracy.
Decoding brain states from functional magnetic resonance imaging (fMRI) data is vital for advancing neuroscience and clinical applications. While traditional machine learning and deep learning approaches have made strides in leveraging the high-dimensional and complex nature of fMRI data, they often fail to utilize the contextual richness provided by Digital Imaging and Communications in Medicine (DICOM) metadata. This paper presents a novel framework integrating transformer-based architectures with multimodal inputs, including fMRI data and DICOM metadata. By employing attention mechanisms, the proposed method captures intricate spatial-temporal patterns and contextual relationships, enhancing model accuracy, interpretability, and robustness. The potential of this framework spans applications in clinical diagnostics, cognitive neuroscience, and personalized medicine. Limitations, such as metadata variability and computational demands, are addressed, and future directions for optimizing scalability and generalizability are discussed.
Medical decision-making makes frequent use of algorithms that combine risk equations with rules, providing clear and standardized treatment pathways. Symbolic regression (SR) traditionally limits its search space to continuous function forms and their parameters, making it difficult to model this decision-making. However, due to its ability to derive data-driven, interpretable models, SR holds promise for developing data-driven clinical risk scores. To that end we introduce Brush, an SR algorithm that combines decision-tree-like splitting algorithms with non-linear constant optimization, allowing for seamless integration of rule-based logic into symbolic regression and classification models. Brush achieves Pareto-optimal performance on SRBench, and was applied to recapitulate two widely used clinical scoring systems, achieving high accuracy and interpretable models. Compared to decision trees, random forests, and other SR methods, Brush achieves comparable or superior predictive performance while producing simpler models.
There are no more papers matching your filters at the moment.