biomolecules
09 Dec 2025
Enzymes are crucial catalysts that enable a wide range of biochemical reactions. Efficiently identifying specific enzymes from vast protein libraries is essential for advancing biocatalysis. Traditional computational methods for enzyme screening and retrieval are time-consuming and resource-intensive. Recently, deep learning approaches have shown promise. However, these methods focus solely on the interaction between enzymes and reactions, overlooking the inherent hierarchical relationships within each domain. To address these limitations, we introduce FGW-CLIP, a novel contrastive learning framework based on optimizing the fused Gromov-Wasserstein distance. FGW-CLIP incorporates multiple alignments, including inter-domain alignment between reactions and enzymes and intra-domain alignment within enzymes and reactions. By introducing a tailored regularization term, our method minimizes the Gromov-Wasserstein distance between enzyme and reaction spaces, which enhances information integration across these domains. Extensive evaluations demonstrate the superiority of FGW-CLIP in challenging enzyme-reaction tasks. On the widely-used EnzymeMap benchmark, FGW-CLIP achieves state-of-the-art performance in enzyme virtual screening, as measured by BEDROC and EF metrics. Moreover, FGW-CLIP consistently outperforms across all three splits of ReactZyme, the largest enzyme-reaction benchmark, demonstrating robust generalization to novel enzymes and reactions. These results position FGW-CLIP as a promising framework for enzyme discovery in complex biochemical settings, with strong adaptability across diverse screening scenarios.
06 Dec 2025
Accurate prediction of protein-protein binding affinity is vital for understanding molecular interactions and designing therapeutics. We adapt Boltz-2, a state-of-the-art structure-based protein-ligand affinity predictor, for protein-protein affinity regression and evaluate it on two datasets, TCR3d and PPB-affinity. Despite high structural accuracy, Boltz-2-PPI underperforms relative to sequence-based alternatives in both small- and larger-scale data regimes. Combining embeddings from Boltz-2-PPI with sequence-based embeddings yields complementary improvements, particularly for weaker sequence models, suggesting different signals are learned by sequence- and structure-based models. Our results echo known biases associated with training with structural data and suggest that current structure-based representations are not primed for performant affinity prediction.
06 Dec 2025
Understanding how protein mutations affect protein structure is essential for advancements in computational biology and bioinformatics. We introduce PRIMRose, a novel approach that predicts energy values for each residue given a mutated protein sequence. Unlike previous models that assess global energy shifts, our method analyzes the localized energetic impact of double amino acid insertions or deletions (InDels) at the individual residue level, enabling residue-specific insights into structural and functional disruption. We implement a Convolutional Neural Network architecture to predict the energy changes of each residue in a protein mutation. We train our model on datasets constructed from nine proteins, grouped into three categories: one set with exhaustive double InDel mutations, another with approximately 145k randomly sampled double InDel mutations, and a third with approximately 80k randomly sampled double InDel mutations. Our model achieves high predictive accuracy across a range of energy metrics as calculated by the Rosetta molecular modeling suite and reveals localized patterns that influence model performance, such as solvent accessibility and secondary structure context. This per-residue analysis offers new insights into the mutational tolerance of specific regions within proteins and provides higher interpretable and biologically meaningful predictions of InDels' effects.
02 Dec 2025
While data augmentation is widely used to train symmetry-agnostic models, it remains unclear how quickly and effectively they learn to respect symmetries. We investigate this by deriving a principled measure of equivariance error that, for convex losses, calculates the percent of total loss attributable to imperfections in learned symmetry. We focus our empirical investigation to 3D-rotation equivariance on high-dimensional molecular tasks (flow matching, force field prediction, denoising voxels) and find that models reduce equivariance error quickly to ≤2\% held-out loss within 1k-10k training steps, a result robust to model and dataset size. This happens because learning 3D-rotational equivariance is an easier learning task, with a smoother and better-conditioned loss landscape, than the main prediction task. For 3D rotations, the loss penalty for non-equivariant models is small throughout training, so they may achieve lower test loss than equivariant models per GPU-hour unless the equivariant ``efficiency gap'' is narrowed. We also experimentally and theoretically investigate the relationships between relative equivariance error, learning gradients, and model parameters.
02 Dec 2025
Accurately predicting protein fitness with minimal experimental data is a persistent challenge in protein engineering. We introduce PRIMO (PRotein In-context Mutation Oracle), a transformer-based framework that leverages in-context learning and test-time training to adapt rapidly to new proteins and assays without large task-specific datasets. By encoding sequence information, auxiliary zero-shot predictions, and sparse experimental labels from many assays as a unified token set in a pre-training masked-language modeling paradigm, PRIMO learns to prioritize promising variants through a preference-based loss function. Across diverse protein families and properties-including both substitution and indel mutations-PRIMO outperforms zero-shot and fully supervised baselines. This work underscores the power of combining large-scale pre-training with efficient test-time adaptation to tackle challenging protein design tasks where data collection is expensive and label availability is limited.
02 Dec 2025
Richman et al. introduce ConforMix, an inference-time algorithm for diffusion models that enhances the sampling of biomolecular conformational landscapes. The method recovers diverse, biologically relevant protein and RNA structures, including experimentally observed states, and enables efficient free energy estimation for rare conformational transitions.
02 Dec 2025
Consistent Synthetic Sequences Unlock Structural Diversity in Fully Atomistic De Novo Protein Design
Consistent Synthetic Sequences Unlock Structural Diversity in Fully Atomistic De Novo Protein Design
High-quality training datasets are crucial for the development of effective protein design models, but existing synthetic datasets often include unfavorable sequence-structure pairs, impairing generative model performance. We leverage ProteinMPNN, whose sequences are experimentally favorable as well as amenable to folding, together with structure prediction models to align high-quality synthetic structures with recoverable synthetic sequences. In that way, we create a new dataset designed specifically for training expressive, fully atomistic protein generators. By retraining La-Proteina, which models discrete residue type and side chain structure in a continuous latent space, on this dataset, we achieve new state-of-the-art results, with improvements of +54% in structural diversity and +27% in co-designability. To validate the broad utility of our approach, we further introduce Proteina Atomistica, a unified flow-based framework that jointly learns the distribution of protein backbone structure, discrete sequences, and atomistic side chains without latent variables. We again find that training on our new sequence-structure data dramatically boosts benchmark performance, improving \method's structural diversity by +73% and co-designability by +5%. Our work highlights the critical importance of aligned sequence-structure data for training high-performance de novo protein design models. All data will be publicly released.
26 Nov 2025
Generative artificial intelligence (AI) models learn probability distributions from data and produce novel samples that capture the salient properties of their training sets. Proteins are particularly attractive for such approaches given their abundant data and the versatility of their representations, ranging from sequences to structures and functions. This versatility has motivated the rapid development of generative models for protein design, enabling the generation of functional proteins and enzymes with unprecedented success. However, because these models mirror their training distribution, they tend to sample from its most probable modes, while low-probability regions, often encoding valuable properties, remain underexplored. To address this challenge, recent work has focused on guiding generative models to produce proteins with user-specified properties, even when such properties are rare or absent from the original training distribution. In this review, we survey and categorize recent advances in conditioning generative models for protein design. We distinguish approaches that modify model parameters, such as reinforcement learning or supervised fine-tuning, from those that keep the model fixed, including conditional generation, retrieval-augmented strategies, Bayesian guidance, and tailored sampling methods. Together, these developments are beginning to enable the steering of generative models toward proteins with desired, and often previously inaccessible, properties.
20 Nov 2025
Reliable evaluation of protein structure predictions remains challenging, as metrics like pLDDT capture energetic stability but often miss subtle errors such as atomic clashes or conformational traps reflecting topological frustration within the protein folding energy landscape. We present CODE (Chain of Diffusion Embeddings), a self evaluating metric empirically found to quantify topological frustration directly from the latent diffusion embeddings of the AlphaFold3 series of structure predictors in a fully unsupervised manner. Integrating this with pLDDT, we propose CONFIDE, a unified evaluation framework that combines energetic and topological perspectives to improve the reliability of AlphaFold3 and related models. CODE strongly correlates with protein folding rates driven by topological frustration, achieving a correlation of 0.82 compared to pLDDT's 0.33 (a relative improvement of 148\%). CONFIDE significantly enhances the reliability of quality evaluation in molecular glue structure prediction benchmarks, achieving a Spearman correlation of 0.73 with RMSD, compared to pLDDT's correlation of 0.42, a relative improvement of 73.8\%. Beyond quality assessment, our approach applies to diverse drug design tasks, including all-atom binder design, enzymatic active site mapping, mutation induced binding affinity prediction, nucleic acid aptamer screening, and flexible protein modeling. By combining data driven embeddings with theoretical insight, CODE and CONFIDE outperform existing metrics across a wide range of biomolecular systems, offering robust and versatile tools to refine structure predictions, advance structural biology, and accelerate drug discovery.
18 Nov 2025
Peptide-based drugs can bind to protein interaction sites that small molecules often cannot, and are easier to produce than large protein drugs. However, designing effective peptide binders is difficult. A typical peptide has an enormous number of possible sequences, and only a few of these will fold into the right 3D shape to match a given protein target. Existing computational methods either generate many candidate sequences without considering how they will fold, or build peptide backbones and then find suitable sequences afterward. Here we introduce ApexGen, a new AI-based framework that simultaneously designs a peptide's amino-acid sequence and its three-dimensional structure to fit a given protein target. For each target, ApexGen produces a full all-atom peptide model in a small number of deterministic integration steps. In tests on hundreds of protein targets, the peptides designed by ApexGen fit tightly onto their target surfaces and cover nearly the entire binding site. These peptides have shapes similar to those found in natural protein-peptide complexes, and they show strong predicted binding affinity in computational experiments. Because ApexGen couples sequence and structure design at every step of Euler integration within a flow-matching sampler, it is much faster and more efficient than prior approaches. This unified method could greatly accelerate the discovery of new peptide-based therapeutics.
28 Oct 2025
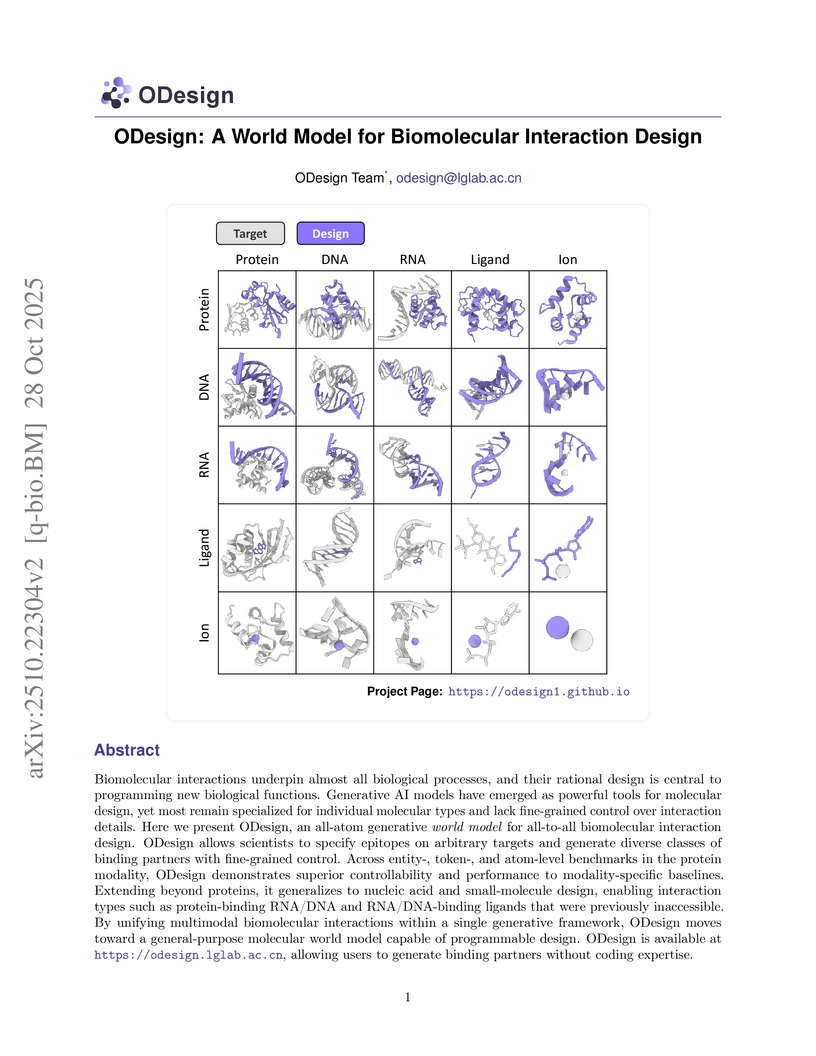
Biomolecular interactions underpin almost all biological processes, and their rational design is central to programming new biological functions. Generative AI models have emerged as powerful tools for molecular design, yet most remain specialized for individual molecular types and lack fine-grained control over interaction details. Here we present ODesign, an all-atom generative world model for all-to-all biomolecular interaction design. ODesign allows scientists to specify epitopes on arbitrary targets and generate diverse classes of binding partners with fine-grained control. Across entity-, token-, and atom-level benchmarks in the protein modality, ODesign demonstrates superior controllability and performance to modality-specific baselines. Extending beyond proteins, it generalizes to nucleic acid and small-molecule design, enabling interaction types such as protein-binding RNA/DNA and RNA/DNA-binding ligands that were previously inaccessible. By unifying multimodal biomolecular interactions within a single generative framework, ODesign moves toward a general-purpose molecular world model capable of programmable design. ODesign is available at this https URL ,
22 Oct 2025
Researchers from the Chinese Academy of Sciences introduce KnowMol, a Molecular Large Language Model that enhances molecular comprehension through a multi-level chemical knowledge dataset (KnowMol-100K) and specialized representation strategies for 1D strings (SELFIES with dedicated vocabulary) and 2D graphs (hierarchical encoder). KnowMol achieves superior performance across molecular understanding and generation tasks, evidenced by a BLEU-4 score of 0.595 in molecule captioning and an Exact Match of 0.752 in forward reaction prediction, while operating with reduced pre-training computational costs.
18 Oct 2025
Applications of machine learning in chemistry are often limited by the scarcity and expense of labeled data, restricting traditional supervised methods. In this work, we introduce a framework for molecular reasoning using general-purpose Large Language Models (LLMs) that operates without requiring labeled training data. Our method anchors chain-of-thought reasoning to the molecular structure by using unique atomic identifiers. First, the LLM performs a one-shot task to identify relevant fragments and their associated chemical labels or transformation classes. In an optional second step, this position-aware information is used in a few-shot task with provided class examples to predict the chemical transformation. We apply our framework to single-step retrosynthesis, a task where LLMs have previously underperformed. Across academic benchmarks and expert-validated drug discovery molecules, our work enables LLMs to achieve high success rates in identifying chemically plausible reaction sites (≥90%), named reaction classes (≥40%), and final reactants (≥74%). Beyond solving complex chemical tasks, our work also provides a method to generate theoretically grounded synthetic datasets by mapping chemical knowledge onto the molecular structure and thereby addressing data scarcity.
20 Oct 2025
A framework standardizes the evaluation of machine-learned molecular dynamics simulations, providing a robust ground-truth dataset, weighted ensemble sampling, and a suite of 19 metrics to rigorously compare models. The framework successfully differentiates model performance and shows ML-MD achieving 10-25x speedups for conformational space exploration.
14 Oct 2025
mRNA design and optimization are important in synthetic biology and therapeutic development, but remain understudied in machine learning. Systematic optimization of mRNAs is hindered by the scarce and imbalanced data as well as complex sequence-function relationships. We present RNAGenScape, a property-guided manifold Langevin dynamics framework that iteratively updates mRNA sequences within a learned latent manifold. RNAGenScape combines an organized autoencoder, which structures the latent space by target properties for efficient and biologically plausible exploration, with a manifold projector that contracts each step of update back to the manifold. RNAGenScape supports property-guided optimization and smooth interpolation between sequences, while remaining robust under scarce and undersampled data, and ensuring that intermediate products are close to the viable mRNA manifold. Across three real mRNA datasets, RNAGenScape improves the target properties with high success rates and efficiency, outperforming various generative or optimization methods developed for proteins or non-biological data. By providing continuous, data-aligned trajectories that reveal how edits influence function, RNAGenScape establishes a scalable paradigm for controllable mRNA design and latent space exploration in mRNA sequence modeling.
13 Oct 2025
Researchers at Wuhan University, Hong Kong Polytechnic University, and Stanford University developed the "Protein-as-Second-Language" framework, enabling general large language models (LLMs) to interpret and reason about protein sequences in a zero-shot setting. This approach, which involves dynamically generating bilingual context from a curated 79,926 QA dataset, achieved an average 7% ROUGE-L improvement and consistently outperformed fine-tuned protein language models on various protein understanding tasks.
29 Sep 2025
Reinforcement learning with stochastic optimal control offers a promising framework for diffusion fine-tuning, where a pre-trained diffusion model is optimized to generate paths that lead to a reward-tilted distribution. While these approaches enable optimization without access to explicit samples from the optimal distribution, they require training on rollouts under the current fine-tuned model, making them susceptible to reinforcing sub-optimal trajectories that yield poor rewards. To overcome this challenge, we introduce TRee Search Guided TRajectory-Aware Fine-Tuning for Discrete Diffusion (TR2-D2), a novel framework that optimizes reward-guided discrete diffusion trajectories with tree search to construct replay buffers for trajectory-aware fine-tuning. These buffers are generated using Monte Carlo Tree Search (MCTS) and subsequently used to fine-tune a pre-trained discrete diffusion model under a stochastic optimal control objective. We validate our framework on single- and multi-objective fine-tuning of biological sequence diffusion models, highlighting the overall effectiveness of TR2-D2 for reliable reward-guided fine-tuning in discrete sequence generation.
05 Sep 2025
Directed evolution is an iterative laboratory process of designing proteins with improved function by iteratively synthesizing new protein variants and evaluating their desired property with expensive and time-consuming biochemical screening. Machine learning methods can help select informative or promising variants for screening to increase their quality and reduce the amount of necessary screening. In this paper, we present a novel method for machine-learning-assisted directed evolution of proteins which combines Bayesian optimization with informative representation of protein variants extracted from a pre-trained protein language model. We demonstrate that the new representation based on the sequence embeddings significantly improves the performance of Bayesian optimization yielding better results with the same number of conducted screening in total. At the same time, our method outperforms the state-of-the-art machine-learning-assisted directed evolution methods with regression objective.
22 Jul 2025
Combinatorial optimization algorithm is essential in computer-aided drug design by progressively exploring chemical space to design lead compounds with high affinity to target protein. However current methods face inherent challenges in integrating domain knowledge, limiting their performance in identifying lead compounds with novel and valid binding mode. Here, we propose AutoLeadDesign, a lead compounds design framework that inspires extensive domain knowledge encoded in large language models with chemical fragments to progressively implement efficient exploration of vast chemical space. The comprehensive experiments indicate that AutoLeadDesign outperforms baseline methods. Significantly, empirical lead design campaigns targeting two clinically relevant targets (PRMT5 and SARS-CoV-2 PLpro) demonstrate AutoLeadDesign's competence in de novo generation of lead compounds achieving expert-competitive design efficacy. Structural analysis further confirms their mechanism-validated inhibitory patterns. By tracing the process of design, we find that AutoLeadDesign shares analogous mechanisms with fragment-based drug design which traditionally rely on the expert decision-making, further revealing why it works. Overall, AutoLeadDesign offers an efficient approach for lead compounds design, suggesting its potential utility in drug design.
01 Jul 2025
Researchers from Princeton and Stanford Universities developed STELLA, a self-evolving large language model agent for biomedical research. This agent leverages a multi-agent architecture and dynamic self-improvement mechanisms to autonomously discover tools and refine its reasoning, achieving state-of-the-art performance on biomedical benchmarks and demonstrating systematic improvement with increased experience.
There are no more papers matching your filters at the moment.