Jülich Research Centre
Recent advancements in measurement techniques have resulted in an increasing amount of data on neural activities recorded in parallel, revealing largely heterogeneous correlation patterns across neurons. Yet, the mechanistic origin of this heterogeneity is largely unknown because existing theoretical approaches linking structure and dynamics in neural circuits are restricted to population-averaged connectivity and activity. Here we present a systematic inclusion of heterogeneity in network connectivity to derive quantitative predictions for neuron-resolved covariances and their statistics in spiking neural networks. Our study shows that the heterogeneity in covariances is not a result of variability in single-neuron firing statistics but stems from the ubiquitously observed sparsity and variability of connections in brain networks. Linear-response theory maps these features to the effective connectivity between neurons, which in turn determines neuronal covariances. Beyond-mean-field tools reveal that synaptic heterogeneity modulates the variability of covariances and thus the complexity of neuronal coordination across many orders of magnitude.
A systematic framework tunes a whole-brain Larter-Breakspear model, using a 998-node human connectome within The Virtual Brain platform, to simultaneously reproduce complex spontaneous and evoked neurophysiological dynamics. The optimized model generates robust alpha rhythms, infra-slow fluctuations, scale-free dynamics, and achieves a Perturbational Complexity Index (PCI) of 0.54, which is significantly higher than the default model's 0.39.
09 Oct 2025
The objective of this paper is to review physiological and computational aspects of the responsiveness of the cerebral cortex to stimulation, and how responsiveness depends on the state of the system. This correspondence between brain state and brain responsiveness (state-dependent responses) is outlined at different scales from the cellular and circuit level, to the mesoscale and macroscale level. At each scale, we review how quantitative methods can be used to characterize network states based on brain responses, such as the Perturbational Complexity Index (PCI). This description will compare data and models, systematically and at multiple scales, with a focus on the mechanisms that explain how brain responses depend on brain states.
Feature learning in neural networks is crucial for their expressive power and inductive biases, motivating various theoretical approaches. Some approaches describe network behavior after training through a change in kernel scale from initialization, resulting in a generalization power comparable to a Gaussian process. Conversely, in other approaches training results in the adaptation of the kernel to the data, involving directional changes to the kernel. The relationship and respective strengths of these two views have so far remained unresolved. This work presents a theoretical framework of multi-scale adaptive feature learning bridging these two views. Using methods from statistical mechanics, we derive analytical expressions for network output statistics which are valid across scaling regimes and in the continuum between them. A systematic expansion of the network's probability distribution reveals that mean-field scaling requires only a saddle-point approximation, while standard scaling necessitates additional correction terms. Remarkably, we find across regimes that kernel adaptation can be reduced to an effective kernel rescaling when predicting the mean network output in the special case of a linear network. However, for linear and non-linear networks, the multi-scale adaptive approach captures directional feature learning effects, providing richer insights than what could be recovered from a rescaling of the kernel alone.
18 Jun 2025
Neuronal activity is found to lie on low-dimensional manifolds embedded within the high-dimensional neuron space. Variants of principal component analysis are frequently employed to assess these manifolds. These methods are, however, limited by assuming a Gaussian data distribution and a flat manifold. In this study, we introduce a method designed to satisfy three core objectives: (1) extract coordinated activity across neurons, described either statistically as correlations or geometrically as manifolds; (2) identify a small number of latent variables capturing these structures; and (3) offer an analytical and interpretable framework characterizing statistical properties by a characteristic function and describing manifold geometry through a collection of charts.
To this end, we employ Normalizing Flows (NFs), which learn an underlying probability distribution of data by an invertible mapping between data and latent space. Their simplicity and ability to compute exact likelihoods distinguish them from other generative networks. We adjust the NF's training objective to distinguish between relevant (in manifold) and noise dimensions (out of manifold). Additionally, we find that different behavioral states align with the components of the latent Gaussian mixture model, enabling their treatment as distinct curved manifolds. Subsequently, we approximate the network for each mixture component with a quadratic mapping, allowing us to characterize both neural manifold curvature and non-Gaussian correlations among recording channels.
Applying the method to recordings in macaque visual cortex, we demonstrate that state-dependent manifolds are curved and exhibit complex statistical dependencies. Our approach thus enables an expressive description of neural population activity, uncovering non-linear interactions among groups of neurons.
Graph neural networks (GNNs) have emerged as powerful tools for processing relational data in applications. However, GNNs suffer from the problem of oversmoothing, the property that the features of all nodes exponentially converge to the same vector over layers, prohibiting the design of deep GNNs. In this work we study oversmoothing in graph convolutional networks (GCNs) by using their Gaussian process (GP) equivalence in the limit of infinitely many hidden features. By generalizing methods from conventional deep neural networks (DNNs), we can describe the distribution of features at the output layer of deep GCNs in terms of a GP: as expected, we find that typical parameter choices from the literature lead to oversmoothing. The theory, however, allows us to identify a new, non-oversmoothing phase: if the initial weights of the network have sufficiently large variance, GCNs do not oversmooth, and node features remain informative even at large depth. We demonstrate the validity of this prediction in finite-size GCNs by training a linear classifier on their output. Moreover, using the linearization of the GCN GP, we generalize the concept of propagation depth of information from DNNs to GCNs. This propagation depth diverges at the transition between the oversmoothing and non-oversmoothing phase. We test the predictions of our approach and find good agreement with finite-size GCNs. Initializing GCNs near the transition to the non-oversmoothing phase, we obtain networks which are both deep and expressive.
The pedagogical review by Ringel et al. (2025) provides a theoretical framework for deep learning using statistical field theory, explaining generalization, implicit bias, and feature learning across various network complexities. It offers derivations for observed scaling laws in large models by moving beyond infinite-width approximations to analyze finite-width effects and their impact on internal representations.
15 Oct 2024
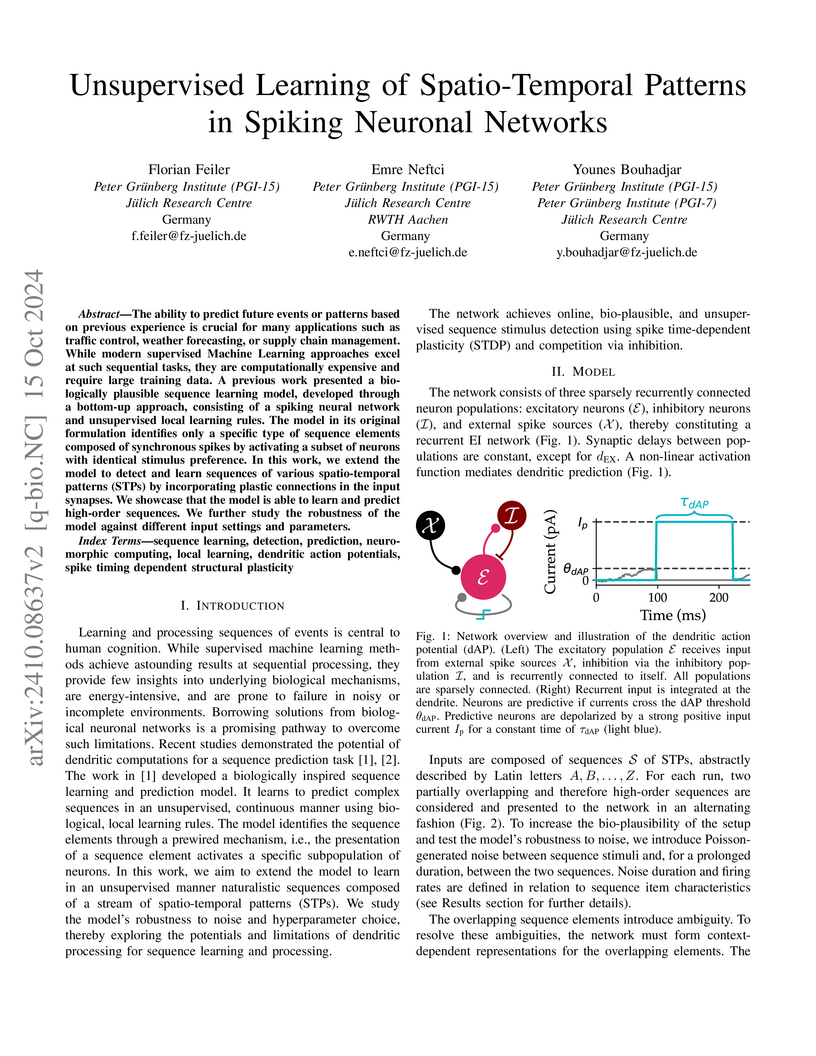
The ability to predict future events or patterns based on previous experience is crucial for many applications such as traffic control, weather forecasting, or supply chain management. While modern supervised Machine Learning approaches excel at such sequential tasks, they are computationally expensive and require large training data. A previous work presented a biologically plausible sequence learning model, developed through a bottom-up approach, consisting of a spiking neural network and unsupervised local learning rules. The model in its original formulation identifies only a specific type of sequence elements composed of synchronous spikes by activating a subset of neurons with identical stimulus preference. In this work, we extend the model to detect and learn sequences of various spatio-temporal patterns (STPs) by incorporating plastic connections in the input synapses. We showcase that the model is able to learn and predict high-order sequences. We further study the robustness of the model against different input settings and parameters.
22 Sep 2017
Researchers at Jülich Research Centre developed an analytical framework for stochastically driven random neural networks, revealing that external input suppresses chaos and that optimal sequential memory occurs in a newly identified 'expansive, non-chaotic' regime, challenging the traditional 'edge of chaos' paradigm. The study provides an exact condition for the onset of chaos in these driven systems and demonstrates that maximal memory is achieved when the network is locally expansive but globally stable.
State-of-the-art neural networks require extreme computational power to
train. It is therefore natural to wonder whether they are optimally trained.
Here we apply a recent advancement in stochastic thermodynamics which allows
bounding the speed at which one can go from the initial weight distribution to
the final distribution of the fully trained network, based on the ratio of
their Wasserstein-2 distance and the entropy production rate of the dynamical
process connecting them. Considering both gradient-flow and Langevin training
dynamics, we provide analytical expressions for these speed limits for linear
and linearizable neural networks e.g. Neural Tangent Kernel (NTK). Remarkably,
given some plausible scaling assumptions on the NTK spectra and spectral
decomposition of the labels -- learning is optimal in a scaling sense. Our
results are consistent with small-scale experiments with Convolutional Neural
Networks (CNNs) and Fully Connected Neural networks (FCNs) on CIFAR-10, showing
a short highly non-optimal regime followed by a longer optimal regime.
Assessing the similarity of matrices is valuable for analyzing the extent to
which data sets exhibit common features in tasks such as data clustering,
dimensionality reduction, pattern recognition, group comparison, and graph
analysis. Methods proposed for comparing vectors, such as cosine similarity,
can be readily generalized to matrices. However, this approach usually neglects
the inherent two-dimensional structure of matrices. Here, we propose singular
angle similarity (SAS), a measure for evaluating the structural similarity
between two arbitrary, real matrices of the same shape based on singular value
decomposition. After introducing the measure, we compare SAS with standard
measures for matrix comparison and show that only SAS captures the
two-dimensional structure of matrices. Further, we characterize the behavior of
SAS in the presence of noise and as a function of matrix dimensionality.
Finally, we apply SAS to two use cases: square non-symmetric matrices of
probabilistic network connectivity, and non-square matrices representing neural
brain activity. For synthetic data of network connectivity, SAS matches
intuitive expectations and allows for a robust assessment of similarities and
differences. For experimental data of brain activity, SAS captures differences
in the structure of high-dimensional responses to different stimuli. We
conclude that SAS is a suitable measure for quantifying the shared structure of
matrices with arbitrary shape.
The majority of research in both training Artificial Neural Networks (ANNs) and modeling learning in biological brains focuses on synaptic plasticity, where learning equates to changing the strength of existing connections. However, in biological brains, structural plasticity - where new connections are created and others removed - is also vital, not only for effective learning but also for recovery from damage and optimal resource usage. Inspired by structural plasticity, pruning is often used in machine learning to remove weak connections from trained models to reduce the computational requirements of inference. However, the machine learning frameworks typically used for backpropagation-based training of both ANNs and Spiking Neural Networks (SNNs) are optimized for dense connectivity, meaning that pruning does not help reduce the training costs of ever-larger models. The GeNN simulator already supports efficient GPU-accelerated simulation of sparse SNNs for computational neuroscience and machine learning. Here, we present a new flexible framework for implementing GPU-accelerated structural plasticity rules and demonstrate this first using the e-prop supervised learning rule and DEEP R to train efficient, sparse SNN classifiers and then, in an unsupervised learning context, to learn topographic maps. Compared to baseline dense models, our sparse classifiers reduce training time by up to 10x while the DEEP R rewiring enables them to perform as well as the original models. We demonstrate topographic map formation in faster-than-realtime simulations, provide insights into the connectivity evolution, and measure simulation speed versus network size. The proposed framework will enable further research into achieving and maintaining sparsity in network structure and neural communication, as well as exploring the computational benefits of sparsity in a range of neuromorphic applications.
To understand how rich dynamics emerge in neural populations, we require
models exhibiting a wide range of activity patterns while remaining
interpretable in terms of connectivity and single-neuron dynamics. However, it
has been challenging to fit such mechanistic spiking networks at the single
neuron scale to empirical population data. To close this gap, we propose to fit
such data at a meso scale, using a mechanistic but low-dimensional and hence
statistically tractable model. The mesoscopic representation is obtained by
approximating a population of neurons as multiple homogeneous `pools' of
neurons, and modelling the dynamics of the aggregate population activity within
each pool. We derive the likelihood of both single-neuron and connectivity
parameters given this activity, which can then be used to either optimize
parameters by gradient ascent on the log-likelihood, or to perform Bayesian
inference using Markov Chain Monte Carlo (MCMC) sampling. We illustrate this
approach using a model of generalized integrate-and-fire neurons for which
mesoscopic dynamics have been previously derived, and show that both
single-neuron and connectivity parameters can be recovered from simulated data.
In particular, our inference method extracts posterior correlations between
model parameters, which define parameter subsets able to reproduce the data. We
compute the Bayesian posterior for combinations of parameters using MCMC
sampling and investigate how the approximations inherent to a mesoscopic
population model impact the accuracy of the inferred single-neuron parameters.
A key property of neural networks driving their success is their ability to learn features from data. Understanding feature learning from a theoretical viewpoint is an emerging field with many open questions. In this work we capture finite-width effects with a systematic theory of network kernels in deep non-linear neural networks. We show that the Bayesian prior of the network can be written in closed form as a superposition of Gaussian processes, whose kernels are distributed with a variance that depends inversely on the network width N . A large deviation approach, which is exact in the proportional limit for the number of data points P=αN→∞, yields a pair of forward-backward equations for the maximum a posteriori kernels in all layers at once. We study their solutions perturbatively to demonstrate how the backward propagation across layers aligns kernels with the target. An alternative field-theoretic formulation shows that kernel adaptation of the Bayesian posterior at finite-width results from fluctuations in the prior: larger fluctuations correspond to a more flexible network prior and thus enable stronger adaptation to data. We thus find a bridge between the classical edge-of-chaos NNGP theory and feature learning, exposing an intricate interplay between criticality, response functions, and feature scale.
Understanding capabilities and limitations of different network architectures is of fundamental importance to machine learning. Bayesian inference on Gaussian processes has proven to be a viable approach for studying recurrent and deep networks in the limit of infinite layer width, n→∞. Here we present a unified and systematic derivation of the mean-field theory for both architectures that starts from first principles by employing established methods from statistical physics of disordered systems. The theory elucidates that while the mean-field equations are different with regard to their temporal structure, they yet yield identical Gaussian kernels when readouts are taken at a single time point or layer, respectively. Bayesian inference applied to classification then predicts identical performance and capabilities for the two architectures. Numerically, we find that convergence towards the mean-field theory is typically slower for recurrent networks than for deep networks and the convergence speed depends non-trivially on the parameters of the weight prior as well as the depth or number of time steps, respectively. Our method exposes that Gaussian processes are but the lowest order of a systematic expansion in 1/n and we compute next-to-leading-order corrections which turn out to be architecture-specific. The formalism thus paves the way to investigate the fundamental differences between recurrent and deep architectures at finite widths n.
03 Sep 2019
The impairment of cognitive function in Alzheimer's is clearly correlated to
synapse loss. However, the mechanisms underlying this correlation are only
poorly understood. Here, we investigate how the loss of excitatory synapses in
sparsely connected random networks of spiking excitatory and inhibitory neurons
alters their dynamical characteristics. Beyond the effects on the network's
activity statistics, we find that the loss of excitatory synapses on excitatory
neurons shifts the network dynamic towards the stable regime. The decrease in
sensitivity to small perturbations to time varying input can be considered as
an indication of a reduction of computational capacity. A full recovery of the
network performance can be achieved by firing rate homeostasis, here
implemented by an up-scaling of the remaining excitatory-excitatory synapses.
By analysing the stability of the linearized network dynamics, we explain how
homeostasis can simultaneously maintain the network's firing rate and
sensitivity to small perturbations.
Simulation speed matters for neuroscientific research: this includes not only how quickly the simulated model time of a large-scale spiking neuronal network progresses, but also how long it takes to instantiate the network model in computer memory. On the hardware side, acceleration via highly parallel GPUs is being increasingly utilized. On the software side, code generation approaches ensure highly optimized code, at the expense of repeated code regeneration and recompilation after modifications to the network model. Aiming for a greater flexibility with respect to iterative model changes, here we propose a new method for creating network connections interactively, dynamically, and directly in GPU memory through a set of commonly used high-level connection rules. We validate the simulation performance with both consumer and data center GPUs on two neuroscientifically relevant models: a cortical microcircuit of about 77,000 leaky-integrate-and-fire neuron models and 300 million static synapses, and a two-population network recurrently connected using a variety of connection rules. With our proposed ad hoc network instantiation, both network construction and simulation times are comparable or shorter than those obtained with other state-of-the-art simulation technologies, while still meeting the flexibility demands of explorative network modeling.
Residual networks have significantly better trainability and thus performance
than feed-forward networks at large depth. Introducing skip connections
facilitates signal propagation to deeper layers. In addition, previous works
found that adding a scaling parameter for the residual branch further improves
generalization performance. While they empirically identified a particularly
beneficial range of values for this scaling parameter, the associated
performance improvement and its universality across network hyperparameters yet
need to be understood. For feed-forward networks, finite-size theories have led
to important insights with regard to signal propagation and hyperparameter
tuning. We here derive a systematic finite-size field theory for residual
networks to study signal propagation and its dependence on the scaling for the
residual branch. We derive analytical expressions for the response function, a
measure for the network's sensitivity to inputs, and show that for deep
networks the empirically found values for the scaling parameter lie within the
range of maximal sensitivity. Furthermore, we obtain an analytical expression
for the optimal scaling parameter that depends only weakly on other network
hyperparameters, such as the weight variance, thereby explaining its
universality across hyperparameters. Overall, this work provides a theoretical
framework to study ResNets at finite size.
07 Nov 2017
Neural-network models of high-level brain functions such as memory recall and
reasoning often rely on the presence of stochasticity. The majority of these
models assumes that each neuron in the functional network is equipped with its
own private source of randomness, often in the form of uncorrelated external
noise. However, both in vivo and in silico, the number of noise sources is
limited due to space and bandwidth constraints. Hence, neurons in large
networks usually need to share noise sources. Here, we show that the resulting
shared-noise correlations can significantly impair the performance of
stochastic network models. We demonstrate that this problem can be overcome by
using deterministic recurrent neural networks as sources of uncorrelated noise,
exploiting the decorrelating effect of inhibitory feedback. Consequently, even
a single recurrent network of a few hundred neurons can serve as a natural
noise source for large ensembles of functional networks, each comprising
thousands of units. We successfully apply the proposed framework to a diverse
set of binary-unit networks with different dimensionalities and entropies, as
well as to a network reproducing handwritten digits with distinct predefined
frequencies. Finally, we show that the same design transfers to functional
networks of spiking neurons.
Neuronal systems need to process temporal signals. We here show how higher-order temporal (co-)fluctuations can be employed to represent and process information. Concretely, we demonstrate that a simple biologically inspired feedforward neuronal model is able to extract information from up to the third order cumulant to perform time series classification. This model relies on a weighted linear summation of synaptic inputs followed by a nonlinear gain function. Training both - the synaptic weights and the nonlinear gain function - exposes how the non-linearity allows for the transfer of higher order correlations to the mean, which in turn enables the synergistic use of information encoded in multiple cumulants to maximize the classification accuracy. The approach is demonstrated both on a synthetic and on real world datasets of multivariate time series. Moreover, we show that the biologically inspired architecture makes better use of the number of trainable parameters as compared to a classical machine-learning scheme. Our findings emphasize the benefit of biological neuronal architectures, paired with dedicated learning algorithms, for the processing of information embedded in higher-order statistical cumulants of temporal (co-)fluctuations.
There are no more papers matching your filters at the moment.