Wellcome Centre for Human NeuroimagingUCL Queen Square Institute of Neurology
17 Sep 2025
Researchers at the University of Oxford and University College London developed the VECTOR framework, leveraging Large Language Models to quantitatively map human thought trajectories from verbal reports into a novel "schema space." This method accurately captures the dynamic organization of conceptual knowledge, predicting behavioral measures and revealing individual differences in communication styles.
10 Jun 2024
Magnetic resonance imaging (MRI) is a crucial tool to identify brain abnormalities in a wide range of neurological disorders. In focal epilepsy MRI is used to identify structural cerebral abnormalities. For covert lesions, machine learning and artificial intelligence algorithms may improve lesion detection if abnormalities are not evident on visual inspection. The success of this approach depends on the volume and quality of training data.
Herein, we release an open-source dataset of preprocessed MRI scans from 442 individuals with drug-refractory focal epilepsy who had neurosurgical resections, and detailed demographic information. The MRI scan data includes the preoperative 3D T1 and where available 3D FLAIR, as well as a manually inspected complete surface reconstruction and volumetric parcellations. Demographic information includes age, sex, age of onset of epilepsy, location of surgery, histopathology of resected specimen, occurrence and frequency of focal seizures with and without impairment of awareness, focal to bilateral tonic-clonic seizures, number of anti-seizure medications (ASMs) at time of surgery, and a total of 1764 patient years of post-surgical follow up. Crucially, we also include resection masks delineated from post-surgical imaging.
To demonstrate the veracity of our data, we successfully replicated previous studies showing long-term outcomes of seizure freedom in the range of around 50%. Our imaging data replicates findings of group level atrophy in patients compared to controls. Resection locations in the cohort were predominantly in the temporal and frontal lobes.
We envisage our dataset, shared openly with the community, will catalyse the development and application of computational methods in clinical neurology.
This research develops a "Bayesian mechanics" for adaptive systems, demonstrating how variational Bayesian inference and active inference naturally emerge in systems possessing a Markov blanket and maintaining a non-equilibrium Gaussian steady-state. It formally unifies statistical physics, information theory, theoretical neuroscience, and stochastic control, showing that expected internal and active states minimize a variational free energy, effectively performing precision-weighted prediction error minimization.
23 Dec 2021
This paper proposes the Insula Hierarchical Modular Adaptive Interoception Control (IMAC) model, a theoretical framework integrating neuroanatomy and active inference to explain the insular cortex's role in interoception, adaptive behavior, and the generation of conscious feelings. The model details how different insula sub-regions process bodily signals to form hierarchical representations, leading to subjective experiences.
 CNRS
CNRS University of Toronto
University of Toronto Imperial College LondonUniversity of Zurich
Imperial College LondonUniversity of Zurich University of Southern CaliforniaUniversity of Bern
University of Southern CaliforniaUniversity of Bern University College London
University College London NVIDIA
NVIDIA KU Leuven
KU Leuven Arizona State UniversitySunnybrook Research Institute
Arizona State UniversitySunnybrook Research Institute King’s College LondonKlinikum rechts der IsarBeijing University of Posts and TelecommunicationsHelmholtz MunichINSA-LyonINSERMTechnical University MunichUniversity of GironaRadboud University Medical CenterPohang University of Science and Technology (POSTECH)Feng Chia UniversityUniversity Medical Center Hamburg-EppendorfUniversity Hospital of ZurichUniversity Hospital BernHelmholtz AICREATISicometrixDeepwise AI LabInselspitalKeck School of MedicineCentre for Medical Image ComputingNational Heart and Lung InstituteWellcome Centre for Human NeuroimagingUniversity Rovira I VirgiliUniversity Institute of Diagnostic and Interventional NeuroradiologyARTORG Center for Biomedical ResearchTranslaTUMStevens Neuroimaging and Informatics InstituteSupport Center of Advanced Neuroimaging (SCAN)Institute for Health SciencesSTADIUS Center for Dynamical Systems, Signal Processing, and Data AnalyticsGraduate School of Artificial Intelligence (GSAI)Institute of Computer Vision and RoboticsLaboratory of Neuro ImagingUniversit ´e Lyon1China Medical University Hsinchu HospitalHurvitz Brain Sciences Research ProgramCenter for Translational Cancer Research
King’s College LondonKlinikum rechts der IsarBeijing University of Posts and TelecommunicationsHelmholtz MunichINSA-LyonINSERMTechnical University MunichUniversity of GironaRadboud University Medical CenterPohang University of Science and Technology (POSTECH)Feng Chia UniversityUniversity Medical Center Hamburg-EppendorfUniversity Hospital of ZurichUniversity Hospital BernHelmholtz AICREATISicometrixDeepwise AI LabInselspitalKeck School of MedicineCentre for Medical Image ComputingNational Heart and Lung InstituteWellcome Centre for Human NeuroimagingUniversity Rovira I VirgiliUniversity Institute of Diagnostic and Interventional NeuroradiologyARTORG Center for Biomedical ResearchTranslaTUMStevens Neuroimaging and Informatics InstituteSupport Center of Advanced Neuroimaging (SCAN)Institute for Health SciencesSTADIUS Center for Dynamical Systems, Signal Processing, and Data AnalyticsGraduate School of Artificial Intelligence (GSAI)Institute of Computer Vision and RoboticsLaboratory of Neuro ImagingUniversit ´e Lyon1China Medical University Hsinchu HospitalHurvitz Brain Sciences Research ProgramCenter for Translational Cancer ResearchDiffusion-weighted MRI (DWI) is essential for stroke diagnosis, treatment decisions, and prognosis. However, image and disease variability hinder the development of generalizable AI algorithms with clinical value. We address this gap by presenting a novel ensemble algorithm derived from the 2022 Ischemic Stroke Lesion Segmentation (ISLES) challenge. ISLES'22 provided 400 patient scans with ischemic stroke from various medical centers, facilitating the development of a wide range of cutting-edge segmentation algorithms by the research community. Through collaboration with leading teams, we combined top-performing algorithms into an ensemble model that overcomes the limitations of individual solutions. Our ensemble model achieved superior ischemic lesion detection and segmentation accuracy on our internal test set compared to individual algorithms. This accuracy generalized well across diverse image and disease variables. Furthermore, the model excelled in extracting clinical biomarkers. Notably, in a Turing-like test, neuroradiologists consistently preferred the algorithm's segmentations over manual expert efforts, highlighting increased comprehensiveness and precision. Validation using a real-world external dataset (N=1686) confirmed the model's generalizability. The algorithm's outputs also demonstrated strong correlations with clinical scores (admission NIHSS and 90-day mRS) on par with or exceeding expert-derived results, underlining its clinical relevance. This study offers two key findings. First, we present an ensemble algorithm (this https URL) that detects and segments ischemic stroke lesions on DWI across diverse scenarios on par with expert (neuro)radiologists. Second, we show the potential for biomedical challenge outputs to extend beyond the challenge's initial objectives, demonstrating their real-world clinical applicability.
One of the primary technical challenges facing magnetoencephalography (MEG) is that the magnitude of neuromagnetic fields is several orders of magnitude lower than interfering signals. Recently, a new type of sensor has been developed - the optically pumped magnetometer (OPM). These sensors can be placed directly on the scalp and move with the head during participant movement, making them wearable. This opens up a range of exciting experimental and clinical opportunities for OPM-based MEG experiments, including paediatric studies, and the incorporation of naturalistic movements into neuroimaging paradigms. However, OPMs face some unique challenges in terms of interference suppression, especially in situations involving mobile participants, and when OPMs are integrated with electrical equipment required for naturalistic paradigms, such as motion capture systems. Here we briefly review various hardware solutions for OPM interference suppression. We then outline several signal processing strategies aimed at increasing the signal from neuromagnetic sources. These include regression-based strategies, temporal filtering and spatial filtering approaches. The focus is on the practical application of these signal processing algorithms to OPM data. In a similar vein, we include two worked-through experiments using OPM data collected from a whole-head sensor array. These tutorial-style examples illustrate how the steps for suppressing external interference can be implemented, including the associated data and code so that researchers can try the pipelines for themselves. With the popularity of OPM-based MEG rising, there will be an increasing need to deal with interference suppression. We hope this practical paper provides a resource for OPM-based MEG researchers to build upon.
13 May 2025
Ultradian rhythms - quasi-rhythmic fluctuations in behavior and physiology
with periods shorter than 24 hours - are observed across various organisms,
including humans. Despite their role in key biological processes such as sleep
architecture and hormone regulation, their underlying mechanisms remain poorly
understood. Here, we leveraged wearable sensor technology for continuous
monitoring of physiological signals in 16 healthy participants over two weeks.
By systematically removing circadian and longer-scale rhythms, we isolated
ultradian dynamics and modeled them using the Hankel Alternative View of
Koopman (HAVOK) framework,a data-driven approach based on Takens' embedding
theorem and Koopman operator theory. This allowed us to characterize ultradian
rhythms as an intermittently forced linear system and distinguish between
regular oscillatory behavior and more complex dynamics. Across participants,
ultradian fluctuations were well-described by the HAVOK model, with
intermittent forcing consistently observed. The model demonstrated strong
forecasting accuracy, with root mean squared error (RMSE) of 0.0315±0.02,
0.0306±0.02, and 0.0218±0.02 in the leading time-delay coordinates.
Notably, a significant sex difference in model rank (z = -2.06, p = 0.0396)
suggests that sex hormones may play a key role in ultradian dynamics. These
findings provide evidence for intermittently forced linear systems as a useful
framework for understanding ultradian rhythms and their regulation.
 University of Cambridge
University of Cambridge University of Southern California
University of Southern California National University of Singapore
National University of Singapore University College London
University College London University of Oxford
University of Oxford Georgia Institute of Technology
Georgia Institute of Technology University of Copenhagen
University of Copenhagen University of California, San Diego
University of California, San Diego McGill University
McGill University Emory University
Emory University University of Pennsylvania
University of Pennsylvania Arizona State University
Arizona State University University of Maryland
University of Maryland King’s College LondonErasmus MCMayo ClinicLund UniversityBrandeis UniversityBen-Gurion University of the NegevUniversity of Eastern FinlandPortland State UniversityUniversity of California San FranciscoNational Institute on AgingGenentechUniversity of PlymouthBanner Alzheimer’s InstituteUCL Queen Square Institute of NeurologyThe University of Texas Health Science Center at HoustonMedical College of WisconsinGerman Center for Neurodegenerative DiseasesUniversity of GhanaInstituto Tecnol ́ogico y de Estudios Superiores de MonterreyH. Lundbeck A/SVU Medical CentreInstitut du Cerveau et de la Moelle ́epini`ereVasile Lucaciu National CollegeBiomarinIBM Research - Australia
King’s College LondonErasmus MCMayo ClinicLund UniversityBrandeis UniversityBen-Gurion University of the NegevUniversity of Eastern FinlandPortland State UniversityUniversity of California San FranciscoNational Institute on AgingGenentechUniversity of PlymouthBanner Alzheimer’s InstituteUCL Queen Square Institute of NeurologyThe University of Texas Health Science Center at HoustonMedical College of WisconsinGerman Center for Neurodegenerative DiseasesUniversity of GhanaInstituto Tecnol ́ogico y de Estudios Superiores de MonterreyH. Lundbeck A/SVU Medical CentreInstitut du Cerveau et de la Moelle ́epini`ereVasile Lucaciu National CollegeBiomarinIBM Research - AustraliaWe present the findings of "The Alzheimer's Disease Prediction Of
Longitudinal Evolution" (TADPOLE) Challenge, which compared the performance of
92 algorithms from 33 international teams at predicting the future trajectory
of 219 individuals at risk of Alzheimer's disease. Challenge participants were
required to make a prediction, for each month of a 5-year future time period,
of three key outcomes: clinical diagnosis, Alzheimer's Disease Assessment Scale
Cognitive Subdomain (ADAS-Cog13), and total volume of the ventricles. The
methods used by challenge participants included multivariate linear regression,
machine learning methods such as support vector machines and deep neural
networks, as well as disease progression models. No single submission was best
at predicting all three outcomes. For clinical diagnosis and ventricle volume
prediction, the best algorithms strongly outperform simple baselines in
predictive ability. However, for ADAS-Cog13 no single submitted prediction
method was significantly better than random guesswork. Two ensemble methods
based on taking the mean and median over all predictions, obtained top scores
on almost all tasks. Better than average performance at diagnosis prediction
was generally associated with the additional inclusion of features from
cerebrospinal fluid (CSF) samples and diffusion tensor imaging (DTI). On the
other hand, better performance at ventricle volume prediction was associated
with inclusion of summary statistics, such as the slope or maxima/minima of
biomarkers. TADPOLE's unique results suggest that current prediction algorithms
provide sufficient accuracy to exploit biomarkers related to clinical diagnosis
and ventricle volume, for cohort refinement in clinical trials for Alzheimer's
disease. However, results call into question the usage of cognitive test scores
for patient selection and as a primary endpoint in clinical trials.
04 Jun 2024
Vagus Nerve Stimulation (VNS) is an established palliative treatment for drug
resistant epilepsy. While effective for many patients, its mechanism of action
is incompletely understood. Predicting individuals' response, or optimum
stimulation parameters, is challenging. Computational modelling has informed
other problems in epilepsy but, to our knowledge, has not been applied to VNS.
We started with an established, four-population neural mass model (NMM),
capable of reproducing the seizure-like dynamics of a thalamocortical circuit.
We extended this to include 18 further neural populations, representing nine
other brain regions relevant to VNS, with connectivity based on existing
literature. We modelled stimulated afferent vagal fibres as projecting to the
nucleus tractus solitarius (NTS), which receives input from the vagus nerve in
vivo.
Bifurcation analysis of a deterministic version of the model showed higher
background NTS input made the model monostable at a fixed point (FP),
representing normal activity, while lower inputs produce bistability between
the FP and a limit cycle (LC), representing the seizure state.
Adding noise produced transitions between seizure and normal states. This
stochastic model spent decreasing time in the seizure state with increasing
background NTS input, until seizures were abolished, consistent with the
deterministic model.
Simulated VNS stimulation, modelled as a 30 Hz square wave, was summed with
the background input to the NTS and was found to reduce total seizure duration
in a dose-dependent manner, similar to expectations in vivo.
We have successfully produced an in silico model of VNS in epilepsy,
capturing behaviour seen in vivo. This may aid understanding therapeutic
mechanisms of VNS in epilepsy and provides a starting point to (i) determine
which patients might respond best to VNS, and (ii) optimise individuals'
treatments.
This paper introduces a novel approach for modelling time-varying connectivity in neuroimaging data, focusing on the slow fluctuations in synaptic efficacy that mediate neuronal dynamics. Building on the framework of Dynamic Causal Modelling (DCM), we propose a method that incorporates temporal basis functions into neural models, allowing for the explicit representation of slow parameter changes. This approach balances expressivity and computational efficiency by modelling these fluctuations as a Gaussian process, offering a middle ground between existing methods that either strongly constrain or excessively relax parameter fluctuations. We validate the ensuing model through simulations and real data from an auditory roving oddball paradigm, demonstrating its potential to explain key aspects of brain dynamics. This work aims to equip researchers with a robust tool for investigating time-varying connectivity, particularly in the context of synaptic modulation and its role in both healthy and pathological brain function.
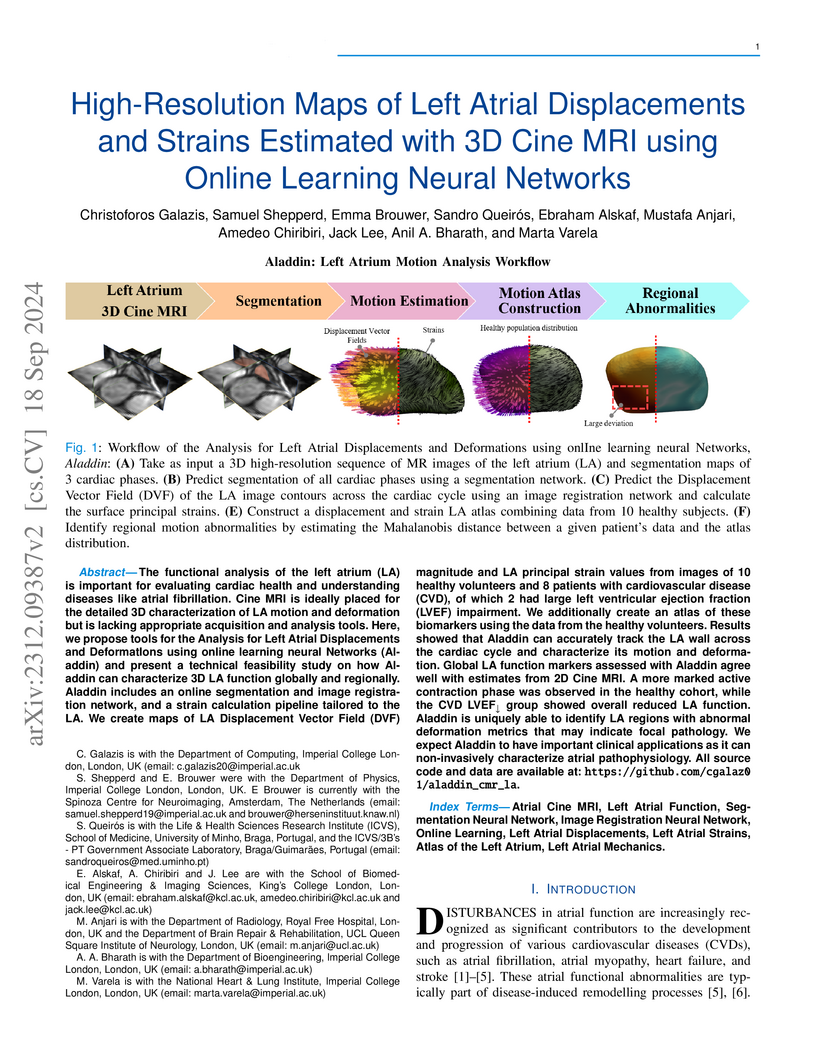
The functional analysis of the left atrium (LA) is important for evaluating cardiac health and understanding diseases like atrial fibrillation. Cine MRI is ideally placed for the detailed 3D characterization of LA motion and deformation but is lacking appropriate acquisition and analysis tools. Here, we propose tools for the Analysis for Left Atrial Displacements and DeformatIons using online learning neural Networks (Aladdin) and present a technical feasibility study on how Aladdin can characterize 3D LA function globally and regionally. Aladdin includes an online segmentation and image registration network, and a strain calculation pipeline tailored to the LA. We create maps of LA Displacement Vector Field (DVF) magnitude and LA principal strain values from images of 10 healthy volunteers and 8 patients with cardiovascular disease (CVD), of which 2 had large left ventricular ejection fraction (LVEF) impairment. We additionally create an atlas of these biomarkers using the data from the healthy volunteers. Results showed that Aladdin can accurately track the LA wall across the cardiac cycle and characterize its motion and deformation. Global LA function markers assessed with Aladdin agree well with estimates from 2D Cine MRI. A more marked active contraction phase was observed in the healthy cohort, while the CVD LVEF group showed overall reduced LA function. Aladdin is uniquely able to identify LA regions with abnormal deformation metrics that may indicate focal pathology. We expect Aladdin to have important clinical applications as it can non-invasively characterize atrial pathophysiology. All source code and data are available at: this https URL.
07 Jan 2022
The identification of abnormal electrographic activity is important in a wide
range of neurological disorders, including epilepsy for localising
epileptogenic tissue. However, this identification may be challenging during
non-seizure (interictal) periods, especially if abnormalities are subtle
compared to the repertoire of possible healthy brain dynamics. Here, we
investigate if such interictal abnormalities become more salient by
quantitatively accounting for the range of healthy brain dynamics in a
location-specific manner. To this end, we constructed a normative map of brain
dynamics, in terms of relative band power, from interictal intracranial
recordings from 234 subjects (21,598 electrode contacts). We then compared
interictal recordings from 62 patients with epilepsy to the normative map to
identify abnormal regions. We hypothesised that if the most abnormal regions
were spared by surgery, then patients would be more likely to experience
continued seizures post-operatively. We first confirmed that the spatial
variations of band power in the normative map across brain regions were
consistent with healthy variations reported in the literature. Second, when
accounting for the normative variations, regions which were spared by surgery
were more abnormal than those resected only in patients with persistent
post-operative seizures (t=-3.6, p=0.0003), confirming our hypothesis. Third,
we found that this effect discriminated patient outcomes (AUC=0.75 p=0.0003).
Normative mapping is a well-established practice in neuroscientific research.
Our study suggests that this approach is feasible to detect interictal
abnormalities in intracranial EEG, and of potential clinical value to identify
pathological tissue in epilepsy. Finally, we make our normative intracranial
map publicly available to facilitate future investigations in epilepsy and
beyond.
27 Feb 2019
This tutorial provides a worked example of using Dynamic Causal Modelling
(DCM) and Parametric Empirical Bayes (PEB) to characterise inter-subject
variability in neural circuitry (effective connectivity). This involves
specifying a hierarchical model with two or more levels. At the first level,
state space models (DCMs) are used to infer the effective connectivity that
best explains a subject's neuroimaging timeseries (e.g. fMRI, MEG, EEG).
Subject-specific connectivity parameters are then taken to the group level,
where they are modelled using a General Linear Model (GLM) that partitions
between-subject variability into designed effects and additive random effects.
The ensuing (Bayesian) hierarchical model conveys both the estimated connection
strengths and their uncertainty (i.e., posterior covariance) from the subject
to the group level; enabling hypotheses to be tested about the commonalities
and differences across subjects. This approach can also finesse parameter
estimation at the subject level, by using the group-level parameters as
empirical priors. We walk through this approach in detail, using data from a
published fMRI experiment that characterised individual differences in
hemispheric lateralization in a semantic processing task. The preliminary
subject specific DCM analysis is covered in detail in a companion paper. This
tutorial is accompanied by the example dataset and step-by-step instructions to
reproduce the analyses.
03 Sep 2024
Anti-seizure medications (ASMs) are the mainstay of treatment for epilepsy, yet their effect on seizure spread is not fully understood. Higher ASM doses have been associated with shorter and less severe seizures. Our objective was to test if this effect was due to limiting seizure spread through early termination of otherwise unchanged seizures.
We retrospectively examined intracranial EEG (iEEG) recordings in 15 subjects that underwent ASM tapering during pre-surgical monitoring. We estimated ASM plasma concentrations based on pharmaco-kinetic modelling. In each subject, we identified seizures that followed the same onset and initial spread patterns, but some seizures terminated early (truncated seizures), and other seizures continued to spread (continuing seizures). We compared ASM concentrations at the times of truncated seizures and continuing seizures.
We found no substantial difference between ASM concentrations when truncated vs. continuing seizures occurred (Mean difference = 4%, sd = 29%, p=0.6).
Our results indicate that ASM did not appear to halt established seizures in this cohort. Further research is needed to understand how ASM may modulate seizure duration and severity.
The analysis of neural power spectra plays a crucial role in understanding brain function and dysfunction. While recent efforts have led to the development of methods for decomposing spectral data, challenges remain in performing statistical analysis and group-level comparisons. Here, we introduce Bayesian Spectral Decomposition (BSD), a Bayesian framework for analysing neural spectral power. BSD allows for the specification, inversion, comparison, and analysis of parametric models of neural spectra, addressing limitations of existing methods. We first establish the face validity of BSD on simulated data and show how it outperforms an established method (\fooof{}) for peak detection on artificial spectral data. We then demonstrate the efficacy of BSD on a group-level study of EEG spectra in 204 healthy subjects from the LEMON dataset. Our results not only highlight the effectiveness of BSD in model selection and parameter estimation, but also illustrate how BSD enables straightforward group-level regression of the effect of continuous covariates such as age. By using Bayesian inference techniques, BSD provides a robust framework for studying neural spectral data and their relationship to brain function and dysfunction.
01 Apr 2025
Objective: The circadian rhythm synchronizes physiological and behavioural patterns with the 24-hour light-dark cycle. Disruption to the circadian rhythm is linked to various health conditions, though optimal methods to describe these disruptions remain unclear. An emerging approach is to examine the intra-individual variability in measurable properties of the circadian rhythm over extended periods. Epileptic seizures are modulated by circadian rhythms, but the relevance of circadian rhythm disruption in epilepsy remains unexplored. Our study investigates intra-individual circadian variability in epilepsy and its relationship with seizures.
Methods: We retrospectively analyzed over 70,000 hours of wearable smartwatch data (Fitbit) from 143 people with epilepsy (PWE) and 31 healthy controls. Circadian oscillations in heart rate time series were extracted, daily estimates of circadian period, acrophase, and amplitude properties were produced, and estimates of the intra-individual variability of these properties over an entire recording were calculated.
Results: PWE exhibited greater intra-individual variability in period (76 min vs. 57 min, d=0.66, p<0.001) and acrophase (64 min vs. 48 min, d=0.49, p=0.004) compared to controls, but not in amplitude (2 bpm, d=-0.15, p=0.49). Variability in circadian properties showed no correlation with seizure frequency, nor any differences between weeks with and without seizures.
Significance: For the first time, we show that heart rate circadian rhythms are more variable in PWE, detectable via consumer wearable devices. However, no association with seizure frequency or occurrence was found, suggesting that this variability might be underpinned by the epilepsy aetiology rather than being a seizure-driven effect.
23 Dec 2024
Anti-seizure medications (ASMs) are the primary treatment for epilepsy, yet medication tapering effects have not been investigated in a dose, region, and time-dependent manner, despite their potential impact on research and clinical practice.
We examined over 3000 hours of intracranial EEG recordings in 32 subjects during long-term monitoring, of which 22 underwent concurrent ASM tapering. We estimated ASM plasma levels based on known pharmaco-kinetics of all the major ASM types.
We found an overall decrease in the power of delta band activity around the period of maximum medication withdrawal in most (80%) subjects, independent of their epilepsy type or medication combination. The degree of withdrawal correlated positively with the magnitude of delta power decrease. This dose-dependent effect was evident across all recorded cortical regions during daytime; but not in sub-cortical regions, or during night time. We found no evidence of a differential effect in seizure onset, spiking, or pathological brain regions.
The finding of decreased delta band power during ASM tapering agrees with previous literature. Our observed dose-dependent effect indicates that monitoring ASM levels in cortical regions may be feasible for applications such as medication reminder systems, or closed-loop ASM delivery systems. ASMs are also used in other neurological and psychiatric conditions, making our findings relevant to a general neuroscience and neurology audience.
Normative models of brain structure estimate the effects of covariates such
as age and sex using large samples of healthy controls. These models can then
be applied to e.g. smaller clinical cohorts to distinguish disease effects from
other covariates. However, these advanced statistical modelling approaches can
be difficult to access, and processing large healthy cohorts is computationally
demanding. Thus, accessible platforms with pre-trained normative models are
needed.
We present such a platform for brain morphology analysis as an open-source
web application this https URL, with six key
features: (i) user-friendly web interface, (ii) individual and group outputs,
(iii) multi-site analysis, (iv) regional and whole-brain analysis, (v)
integration with existing tools, and (vi) featuring multiple morphology
metrics.
Using a diverse sample of 3,276 healthy controls across 21 sites, we
pre-trained normative models on various metrics. We validated the models with a
small sample of individuals with bipolar disorder, showing outputs that aligned
closely with existing literature only after applying our normative modelling.
Using a cohort of people with temporal lobe epilepsy, we showed that
individual-level abnormalities were in line with seizure lateralisation.
Finally, with the ability to investigate multiple morphology measures in the
same framework, we found that biological covariates are better explained in
specific morphology measures, and for applications, only some measures are
sensitive to the disease process.
Our platform offers a comprehensive framework to analyse brain morphology in
clinical and research settings. Validations confirm the superiority of
normative models and the advantage of investigating a range of brain morphology
metrics together.
06 Feb 2025
Normative mapping is a framework used to map population-level features of
health-related variables. It is widely used in neuroscience research, but the
literature lacks established protocols in modalities that do not support
healthy control measurements, such as intracranial EEG (icEEG). An icEEG
normative map would allow researchers to learn about population-level brain
activity and enable comparison of individual data against these norms to
identify abnormalities. Currently, no standardised guide exists for
transforming clinical data into a normative, regional icEEG map. Papers often
cite different software and numerous articles to summarise the lengthy method,
making it laborious for other researchers to understand or apply the process.
Our protocol seeks to remedy this gap by providing a dataflow guide and key
decision points that summarise existing methods. This protocol is used heavily
in published works from our own lab (twelve peer-reviewed journal
publications). Briefly, we take as input, icEEG recordings and neuroimaging
data from people with epilepsy who are undergoing evaluation for resective
surgery. As final outputs, we obtain a normative icEEG map, comprising signal
properties localised to brain regions. Optionally, we can also process new
subjects through the same pipeline and obtain their z-scores (or centiles) in
each brain region, for abnormality detection and localisation. To date, a
single, cohesive, dataflow pipeline for generating normative icEEG maps, along
with abnormality mapping, has not been created. We envisage that this dataflow
guide will not only increase understanding and application of normative mapping
methods, but will also improve the consistency and quality of studies in the
field.
24 Feb 2025
MRI-based delineation of brain tissue removed by epilepsy surgery can be
challenging due to post-operative brain shift. In consequence, most studies use
manual approaches which are prohibitively time-consuming for large sample
sizes, require expertise, and can be prone to errors.
We propose RAMPS (Resections And Masks in Preoperative Space), an automated
pipeline to generate a 3D resection mask of pre-operative tissue. Our pipeline
leverages existing software including FreeSurfer, SynthStrip, Sythnseg and ANTS
to generate a mask in the same space as the patient's pre-operative T1 weighted
MRI. We compare our automated masks against manually drawn masks and two other
existing pipelines (Epic-CHOP and ResectVol).
Comparing to manual masks (N=87), RAMPS achieved a median(IQR) dice
similarity of 0.86(0.078) in temporal lobe resections, and 0.72(0.32) in
extratemporal resections. In comparison to other pipelines, RAMPS had higher
dice similarities (N=62) (RAMPS:0.86, Epic-CHOP: 0.72, ResectVol: 0.72).
We release a user-friendly, easy to use pipeline, RAMPS, open source for
accurate delineation of resected tissue.
There are no more papers matching your filters at the moment.