Siemens Healthcare GmbH
Denoising of clinical CT images is an active area for deep learning research.
Current clinically approved methods use iterative reconstruction methods to
reduce the noise in CT images. Iterative reconstruction techniques require
multiple forward and backward projections, which are time-consuming and
computationally expensive. Recently, deep learning methods have been
successfully used to denoise CT images. However, conventional deep learning
methods suffer from the 'black box' problem. They have low accountability,
which is necessary for use in clinical imaging situations. In this paper, we
use a Joint Bilateral Filter (JBF) to denoise our CT images. The guidance image
of the JBF is estimated using a deep residual convolutional neural network
(CNN). The range smoothing and spatial smoothing parameters of the JBF are
tuned by a deep reinforcement learning task. Our actor first chooses a
parameter, and subsequently chooses an action to tune the value of the
parameter. A reward network is designed to direct the reinforcement learning
task. Our denoising method demonstrates good denoising performance, while
retaining structural information. Our method significantly outperforms state of
the art deep neural networks. Moreover, our method has only two parameters,
which makes it significantly more interpretable and reduces the 'black box'
problem. We experimentally measure the impact of our intelligent parameter
optimization and our reward network. Our studies show that our current setup
yields the best results in terms of structural preservation.
Incorporating computed tomography (CT) reconstruction operators into
differentiable pipelines has proven beneficial in many applications. Such
approaches usually focus on the projection data and keep the acquisition
geometry fixed. However, precise knowledge of the acquisition geometry is
essential for high quality reconstruction results. In this paper, the
differentiable formulation of fan-beam CT reconstruction is extended to the
acquisition geometry. This allows to propagate gradient information from a loss
function on the reconstructed image into the geometry parameters. As a
proof-of-concept experiment, this idea is applied to rigid motion compensation.
The cost function is parameterized by a trained neural network which regresses
an image quality metric from the motion affected reconstruction alone. Using
the proposed method, we are the first to optimize such an autofocus-inspired
algorithm based on analytical gradients. The algorithm achieves a reduction in
MSE by 35.5 % and an improvement in SSIM by 12.6 % over the motion affected
reconstruction. Next to motion compensation, we see further use cases of our
differentiable method for scanner calibration or hybrid techniques employing
deep models.
X-ray based measurement and guidance are commonly used tools in orthopaedic surgery to facilitate a minimally invasive workflow. Typically, a surgical planning is first performed using knowledge of bone morphology and anatomical landmarks. Information about bone location then serves as a prior for registration during overlay of the planning on intra-operative X-ray images. Performing these steps manually however is prone to intra-rater/inter-rater variability and increases task complexity for the surgeon. To remedy these issues, we propose an automatic framework for planning and subsequent overlay. We evaluate it on the example of femoral drill site planning for medial patellofemoral ligament reconstruction surgery. A deep multi-task stacked hourglass network is trained on 149 conventional lateral X-ray images to jointly localize two femoral landmarks, to predict a region of interest for the posterior femoral cortex tangent line, and to perform semantic segmentation of the femur, patella, tibia, and fibula with adaptive task complexity weighting. On 38 clinical test images the framework achieves a median localization error of 1.50 mm for the femoral drill site and mean IOU scores of 0.99, 0.97, 0.98, and 0.96 for the femur, patella, tibia, and fibula respectively. The demonstrated approach consistently performs surgical planning at expert-level precision without the need for manual correction.
Coronary CT angiography (CCTA) has established its role as a non-invasive
modality for the diagnosis of coronary artery disease (CAD). The CAD-Reporting
and Data System (CAD-RADS) has been developed to standardize communication and
aid in decision making based on CCTA findings. The CAD-RADS score is determined
by manual assessment of all coronary vessels and the grading of lesions within
the coronary artery tree.
We propose a bottom-up approach for fully-automated prediction of this score
using deep-learning operating on a segment-wise representation of the coronary
arteries. The method relies solely on a prior fully-automated centerline
extraction and segment labeling and predicts the segment-wise stenosis degree
and the overall calcification grade as auxiliary tasks in a multi-task learning
setup.
We evaluate our approach on a data collection consisting of 2,867 patients.
On the task of identifying patients with a CAD-RADS score indicating the need
for further invasive investigation our approach reaches an area under curve
(AUC) of 0.923 and an AUC of 0.914 for determining whether the patient suffers
from CAD. This level of performance enables our approach to be used in a
fully-automated screening setup or to assist diagnostic CCTA reading,
especially due to its neural architecture design -- which allows comprehensive
predictions.
The positive outcome of a trauma intervention depends on an intraoperative
evaluation of inserted metallic implants. Due to occurring metal artifacts, the
quality of this evaluation heavily depends on the performance of so-called
Metal Artifact Reduction methods (MAR). The majority of these MAR methods
require prior segmentation of the inserted metal objects. Therefore, typically
a rather simple thresholding-based segmentation method in the reconstructed 3D
volume is applied, despite some major disadvantages. With this publication, the
potential of shifting the segmentation task to a learning-based,
view-consistent 2D projection-based method on the downstream MAR's outcome is
investigated. For segmenting the present metal, a rather simple learning-based
2D projection-wise segmentation network that is trained using real data
acquired during cadaver studies, is examined. To overcome the disadvantages
that come along with a 2D projection-wise segmentation, a Consistency Filter is
proposed. The influence of the shifted segmentation domain is investigated by
comparing the results of the standard fsMAR with a modified fsMAR version using
the new segmentation masks. With a quantitative and qualitative evaluation on
real cadaver data, the investigated approach showed an increased MAR
performance and a high insensitivity against metal artifacts. For cases with
metal outside the reconstruction's FoV or cases with vanishing metal, a
significant reduction in artifacts could be shown. Thus, increases of up to
roughly 3 dB w.r.t. the mean PSNR metric over all slices and up to 9 dB for
single slices were achieved. The shown results reveal a beneficial influence of
the shift to a 2D-based segmentation method on real data for downstream use
with a MAR method, like the fsMAR.
Robustness of deep learning methods for limited angle tomography is
challenged by two major factors: a) due to insufficient training data the
network may not generalize well to unseen data; b) deep learning methods are
sensitive to noise. Thus, generating reconstructed images directly from a
neural network appears inadequate. We propose to constrain the reconstructed
images to be consistent with the measured projection data, while the unmeasured
information is complemented by learning based methods. For this purpose, a data
consistent artifact reduction (DCAR) method is introduced: First, a prior image
is generated from an initial limited angle reconstruction via deep learning as
a substitute for missing information. Afterwards, a conventional iterative
reconstruction algorithm is applied, integrating the data consistency in the
measured angular range and the prior information in the missing angular range.
This ensures data integrity in the measured area, while inaccuracies
incorporated by the deep learning prior lie only in areas where no information
is acquired. The proposed DCAR method achieves significant image quality
improvement: for 120-degree cone-beam limited angle tomography more than 10%
RMSE reduction in noise-free case and more than 24% RMSE reduction in noisy
case compared with a state-of-the-art U-Net based method.
Chronic obstructive pulmonary disease (COPD) is a lung disease that is not
fully reversible and one of the leading causes of morbidity and mortality in
the world. Early detection and diagnosis of COPD can increase the survival rate
and reduce the risk of COPD progression in patients. Currently, the primary
examination tool to diagnose COPD is spirometry. However, computed tomography
(CT) is used for detecting symptoms and sub-type classification of COPD. Using
different imaging modalities is a difficult and tedious task even for
physicians and is subjective to inter-and intra-observer variations. Hence,
developing meth-ods that can automatically classify COPD versus healthy
patients is of great interest. In this paper, we propose a 3D deep learning
approach to classify COPD and emphysema using volume-wise annotations only. We
also demonstrate the impact of transfer learning on the classification of
emphysema using knowledge transfer from a pre-trained COPD classification
model.
11 Sep 2017
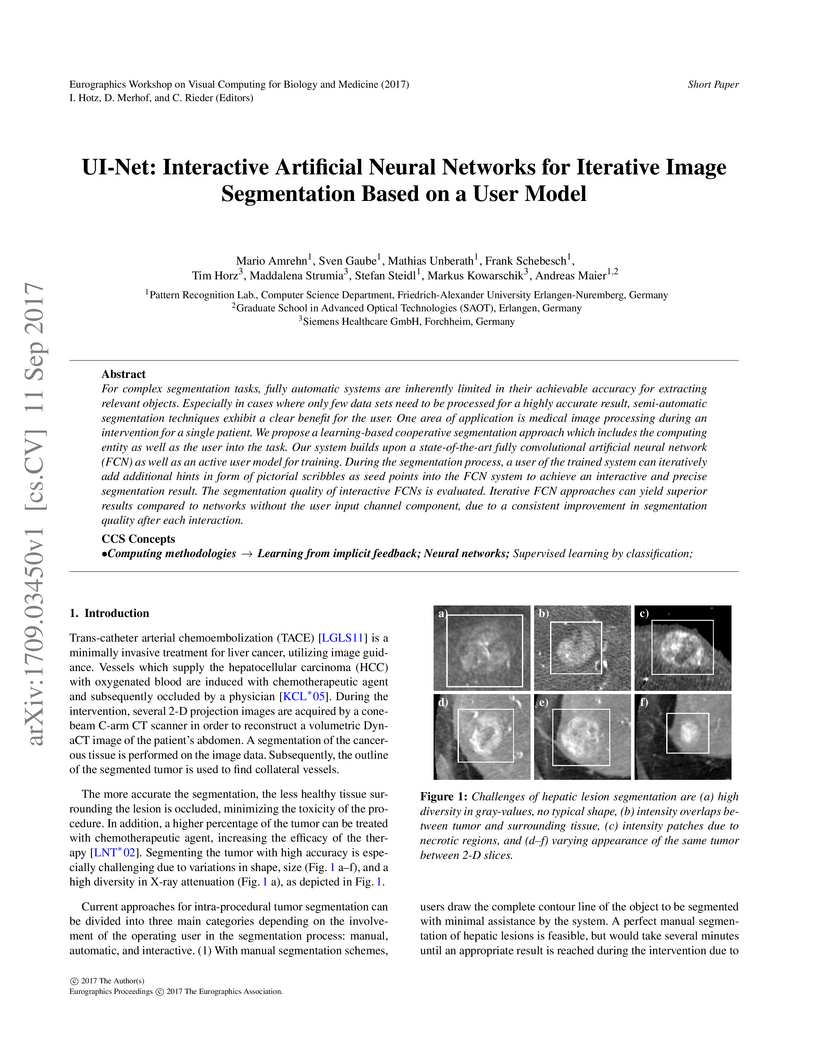
For complex segmentation tasks, fully automatic systems are inherently
limited in their achievable accuracy for extracting relevant objects.
Especially in cases where only few data sets need to be processed for a highly
accurate result, semi-automatic segmentation techniques exhibit a clear benefit
for the user. One area of application is medical image processing during an
intervention for a single patient. We propose a learning-based cooperative
segmentation approach which includes the computing entity as well as the user
into the task. Our system builds upon a state-of-the-art fully convolutional
artificial neural network (FCN) as well as an active user model for training.
During the segmentation process, a user of the trained system can iteratively
add additional hints in form of pictorial scribbles as seed points into the FCN
system to achieve an interactive and precise segmentation result. The
segmentation quality of interactive FCNs is evaluated. Iterative FCN approaches
can yield superior results compared to networks without the user input channel
component, due to a consistent improvement in segmentation quality after each
interaction.
Ultrasound b-mode imaging is a qualitative approach and diagnostic quality
strongly depends on operators' training and experience. Quantitative approaches
can provide information about tissue properties; therefore, can be used for
identifying various tissue types, e.g., speed-of-sound in the tissue can be
used as a biomarker for tissue malignancy, especially in breast imaging. Recent
studies showed the possibility of speed-of-sound reconstruction using deep
neural networks that are fully trained on simulated data. However, because of
the ever-present domain shift between simulated and measured data, the
stability and performance of these models in real setups are still under
debate. In prior works, for training data generation, tissue structures were
modeled as simplified geometrical structures which does not reflect the
complexity of the real tissues. In this study, we proposed a new simulation
setup for training data generation based on Tomosynthesis images. We combined
our approach with the simplified geometrical model and investigated the impacts
of training data diversity on the stability and robustness of an existing
network architecture. We studied the sensitivity of the trained network to
different simulation parameters, e.g., echogenicity, number of scatterers,
noise, and geometry. We showed that the network trained with the joint set of
data is more stable on out-of-domain simulated data as well as measured phantom
data.
With coronary artery disease (CAD) persisting to be one of the leading causes
of death worldwide, interest in supporting physicians with algorithms to speed
up and improve diagnosis is high. In clinical practice, the severeness of CAD
is often assessed with a coronary CT angiography (CCTA) scan and manually
graded with the CAD-Reporting and Data System (CAD-RADS) score. The clinical
questions this score assesses are whether patients have CAD or not (rule-out)
and whether they have severe CAD or not (hold-out). In this work, we reach new
state-of-the-art performance for automatic CAD-RADS scoring. We propose using
severity-based label encoding, test time augmentation (TTA) and model
ensembling for a task-specific deep learning architecture. Furthermore, we
introduce a novel task- and model-specific, heuristic coronary segment
labeling, which subdivides coronary trees into consistent parts across
patients. It is fast, robust, and easy to implement. We were able to raise the
previously reported area under the receiver operating characteristic curve
(AUC) from 0.914 to 0.942 in the rule-out and from 0.921 to 0.950 in the
hold-out task respectively.
Self-supervised image denoising techniques emerged as convenient methods that allow training denoising models without requiring ground-truth noise-free data. Existing methods usually optimize loss metrics that are calculated from multiple noisy realizations of similar images, e.g., from neighboring tomographic slices. However, those approaches fail to utilize the multiple contrasts that are routinely acquired in medical imaging modalities like MRI or dual-energy CT. In this work, we propose the new self-supervised training scheme Noise2Contrast that combines information from multiple measured image contrasts to train a denoising model. We stack denoising with domain-transfer operators to utilize the independent noise realizations of different image contrasts to derive a self-supervised loss. The trained denoising operator achieves convincing quantitative and qualitative results, outperforming state-of-the-art self-supervised methods by 4.7-11.0%/4.8-7.3% (PSNR/SSIM) on brain MRI data and by 43.6-50.5%/57.1-77.1% (PSNR/SSIM) on dual-energy CT X-ray microscopy data with respect to the noisy baseline. Our experiments on different real measured data sets indicate that Noise2Contrast training generalizes to other multi-contrast imaging modalities.
Patient-specific hemodynamics assessment could support diagnosis and
treatment of neurovascular diseases. Currently, conventional medical imaging
modalities are not able to accurately acquire high-resolution hemodynamic
information that would be required to assess complex neurovascular pathologies.
Therefore, computational fluid dynamics (CFD) simulations can be applied to
tomographic reconstructions to obtain clinically relevant information. However,
three-dimensional (3D) CFD simulations require enormous computational resources
and simulation-related expert knowledge that are usually not available in
clinical environments. Recently, deep-learning-based methods have been proposed
as CFD surrogates to improve computational efficiency. Nevertheless, the
prediction of high-resolution transient CFD simulations for complex vascular
geometries poses a challenge to conventional deep learning models. In this
work, we present an architecture that is tailored to predict high-resolution
(spatial and temporal) velocity fields for complex synthetic vascular
geometries. For this, an octree-based spatial discretization is combined with
an implicit neural function representation to efficiently handle the prediction
of the 3D velocity field for each time step. The presented method is evaluated
for the task of cerebral hemodynamics prediction before and during the
injection of contrast agent in the internal carotid artery (ICA). Compared to
CFD simulations, the velocity field can be estimated with a mean absolute error
of 0.024 m/s, whereas the run time reduces from several hours on a
high-performance cluster to a few seconds on a consumer graphical processing
unit.
Objectives Parametric tissue mapping enables quantitative cardiac tissue characterization but is limited by inter-observer variability during manual delineation. Traditional approaches relying on average relaxation values and single cutoffs may oversimplify myocardial complexity. This study evaluates whether deep learning (DL) can achieve segmentation accuracy comparable to inter-observer variability, explores the utility of statistical features beyond mean T1/T2 values, and assesses whether machine learning (ML) combining multiple features enhances disease detection. Materials & Methods T1 and T2 maps were manually segmented. The test subset was independently annotated by two observers, and inter-observer variability was assessed. A DL model was trained to segment left ventricle blood pool and myocardium. Average (A), lower quartile (LQ), median (M), and upper quartile (UQ) were computed for the myocardial pixels and employed in classification by applying cutoffs or in ML. Dice similarity coefficient (DICE) and mean absolute percentage error evaluated segmentation performance. Bland-Altman plots assessed inter-user and model-observer agreement. Receiver operating characteristic analysis determined optimal cutoffs. Pearson correlation compared features from model and manual segmentations. F1-score, precision, and recall evaluated classification performance. Wilcoxon test assessed differences between classification methods, with p < 0.05 considered statistically significant. Results 144 subjects were split into training (100), validation (15) and evaluation (29) subsets. Segmentation model achieved a DICE of 85.4%, surpassing inter-observer agreement. Random forest applied to all features increased F1-score (92.7%, p < 0.001). Conclusion DL facilitates segmentation of T1/ T2 maps. Combining multiple features with ML improves disease detection.
We propose Nonlinear Dipole Inversion (NDI) for high-quality Quantitative
Susceptibility Mapping (QSM) without regularization tuning, while matching the
image quality of state-of-the-art reconstruction techniques. In addition to
avoiding over-smoothing that these techniques often suffer from, we also
obviate the need for parameter selection. NDI is flexible enough to allow for
reconstruction from an arbitrary number of head orientations, and outperforms
COSMOS even when using as few as 1-direction data. This is made possible by a
nonlinear forward-model that uses the magnitude as an effective prior, for
which we derived a simple gradient descent update rule. We synergistically
combine this physics-model with a Variational Network (VN) to leverage the
power of deep learning in the VaNDI algorithm. This technique adopts the simple
gradient descent rule from NDI and learns the network parameters during
training, hence requires no additional parameter tuning. Further, we evaluate
NDI at 7T using highly accelerated Wave-CAIPI acquisitions at 0.5 mm isotropic
resolution and demonstrate high-quality QSM from as few as 2-direction data.
We propose a novel point cloud based 3D organ segmentation pipeline utilizing
deep Q-learning. In order to preserve shape properties, the learning process is
guided using a statistical shape model. The trained agent directly predicts
piece-wise linear transformations for all vertices in each iteration. This
mapping between the ideal transformation for an object outline estimation is
learned based on image features. To this end, we introduce aperture features
that extract gray values by sampling the 3D volume within the cone centered
around the associated vertex and its normal vector. Our approach is also
capable of estimating a hierarchical pyramid of non rigid deformations for
multi-resolution meshes. In the application phase, we use a marginal approach
to gradually estimate affine as well as non-rigid transformations. We performed
extensive evaluations to highlight the robust performance of our approach on a
variety of challenge data as well as clinical data. Additionally, our method
has a run time ranging from 0.3 to 2.7 seconds to segment each organ. In
addition, we show that the proposed method can be applied to different organs,
X-ray based modalities, and scanning protocols without the need of transfer
learning. As we learn actions, even unseen reference meshes can be processed as
demonstrated in an example with the Visible Human. From this we conclude that
our method is robust, and we believe that our method can be successfully
applied to many more applications, in particular, in the interventional imaging
space.
Purpose: To develop an algorithm for the retrospective correction of signal dropout artifacts in abdominal diffusion-weighted imaging (DWI) resulting from cardiac motion.
Methods: Given a set of image repetitions for a slice, a locally adaptive weighted averaging is proposed which aims to suppress the contribution of image regions affected by signal dropouts. Corresponding weight maps were estimated by a sliding-window algorithm which analyzed signal deviations from a patch-wise reference. In order to ensure the computation of a robust reference, repetitions were filtered by a classifier that was trained to detect images corrupted by signal dropouts. The proposed method, termed Deep Learning-guided Adaptive Weighted Averaging (DLAWA), was evaluated in terms of dropout suppression capability, bias reduction in the Apparent Diffusion Coefficient (ADC) and noise characteristics.
Results: In the case of uniform averaging, motion-related dropouts caused signal attenuation and ADC overestimation in parts of the liver with the left lobe being affected particularly. Both effects could be substantially mitigated by DLAWA while preventing global penalties with respect to signal-to-noise ratio (SNR) due to local signal suppression. Performing evaluations on patient data, the capability to recover lesions concealed by signal dropouts was demonstrated as well. Further, DLAWA allowed for transparent control of the trade-off between SNR and signal dropout suppression by means of a few hyperparameters.
Conclusion: This work presents an effective and flexible method for the local compensation of signal dropouts resulting from motion and pulsation. Since DLAWA follows a retrospective approach, no changes to the acquisition are required.
Ischemic strokes are often caused by large vessel occlusions (LVOs), which
can be visualized and diagnosed with Computed Tomography Angiography scans. As
time is brain, a fast, accurate and automated diagnosis of these scans is
desirable. Human readers compare the left and right hemispheres in their
assessment of strokes. A large training data set is required for a standard
deep learning-based model to learn this strategy from data. As labeled medical
data in this field is rare, other approaches need to be developed. To both
include the prior knowledge of side comparison and increase the amount of
training data, we propose an augmentation method that generates artificial
training samples by recombining vessel tree segmentations of the hemispheres or
hemisphere subregions from different patients. The subregions cover vessels
commonly affected by LVOs, namely the internal carotid artery (ICA) and middle
cerebral artery (MCA). In line with the augmentation scheme, we use a
3D-DenseNet fed with task-specific input, fostering a side-by-side comparison
between the hemispheres. Furthermore, we propose an extension of that
architecture to process the individual hemisphere subregions. All
configurations predict the presence of an LVO, its side, and the affected
subregion. We show the effect of recombination as an augmentation strategy in a
5-fold cross validated ablation study. We enhanced the AUC for patient-wise
classification regarding the presence of an LVO of all investigated
architectures. For one variant, the proposed method improved the AUC from 0.73
without augmentation to 0.89. The best configuration detects LVOs with an AUC
of 0.91, LVOs in the ICA with an AUC of 0.96, and in the MCA with 0.91 while
accurately predicting the affected side.
In this paper, we propose a method for denoising diffusion-weighted images
(DWI) of the brain using a convolutional neural network trained on realistic,
synthetic MR data. We compare our results to averaging of repeated scans, a
widespread method used in clinics to improve signal-to-noise ratio of MR
images. To obtain training data for transfer learning, we model, in a
data-driven fashion, the effects of echo-planar imaging (EPI): Nyquist ghosting
and ramp sampling. We introduce these effects to the digital phantom of brain
anatomy (BrainWeb). Instead of simulating pseudo-random noise with a defined
probability distribution, we perform noise scans with a brain-DWI-designed
protocol to obtain realistic noise maps. We combine them with the simulated,
noise-free EPI images. We also measure the Point Spread Function in a DW image
of an AJR-approved geometrical phantom and inter-scan movement in a brain scan
of a healthy volunteer. Their influence on image denoising and averaging of
repeated images is investigated at different signal-to-noise ratio levels.
Denoising performance is evaluated quantitatively using the simulated EPI
images and qualitatively in real EPI DWI of the brain. We show that the
application of our method allows for a significant reduction in scan time by
lowering the number of repeated scans. Visual comparisons made in the acquired
brain images indicate that the denoised single-repetition images are less noisy
than multi-repetition averaged images. We also analyse the convolutional neural
network denoiser and point out the challenges accompanying this denoising
method.
Quantitative ultrasound, e.g., speed-of-sound (SoS) in tissues, provides
information about tissue properties that have diagnostic value. Recent studies
showed the possibility of extracting SoS information from pulse-echo ultrasound
raw data (a.k.a. RF data) using deep neural networks that are fully trained on
simulated data. These methods take sensor domain data, i.e., RF data, as input
and train a network in an end-to-end fashion to learn the implicit mapping
between the RF data domain and SoS domain. However, such networks are prone to
overfitting to simulated data which results in poor performance and instability
when tested on measured data. We propose a novel method for SoS mapping
employing learned representations from two linked autoencoders. We test our
approach on simulated and measured data acquired from human breast mimicking
phantoms. We show that SoS mapping is possible using linked autoencoders. The
proposed method has a Mean Absolute Percentage Error (MAPE) of 2.39% on the
simulated data. On the measured data, the predictions of the proposed method
are close to the expected values with MAPE of 1.1%. Compared to an end-to-end
trained network, the proposed method shows higher stability and
reproducibility.
The anatomical axis of long bones is an important reference line for guiding
fracture reduction and assisting in the correct placement of guide pins,
screws, and implants in orthopedics and trauma surgery. This study investigates
an automatic approach for detection of such axes on X-ray images based on the
segmentation contour of the bone. For this purpose, we use the medically
established two-line method and translate it into a learning-based approach.
The proposed method is evaluated on 38 clinical test images of the femoral and
tibial bone and achieves a median angulation error of 0.19{\deg} and 0.33{\deg}
respectively. An inter-rater study with three trauma surgery experts confirms
reliability of the method and recommends further clinical application.
There are no more papers matching your filters at the moment.