Sunnybrook Research Institute
Current self-supervised learning methods for 3D medical imaging rely on simple pretext formulations and organ- or modality-specific datasets, limiting their generalizability and scalability. We present 3DINO, a cutting-edge SSL method adapted to 3D datasets, and use it to pretrain 3DINO-ViT: a general-purpose medical imaging model, on an exceptionally large, multimodal, and multi-organ dataset of ~100,000 3D medical imaging scans from over 10 organs. We validate 3DINO-ViT using extensive experiments on numerous medical imaging segmentation and classification tasks. Our results demonstrate that 3DINO-ViT generalizes across modalities and organs, including out-of-distribution tasks and datasets, outperforming state-of-the-art methods on the majority of evaluation metrics and labeled dataset sizes. Our 3DINO framework and 3DINO-ViT will be made available to enable research on 3D foundation models or further finetuning for a wide range of medical imaging applications.
Prediction tasks in digital pathology are challenging due to the massive size of whole-slide images (WSIs) and the weak nature of training signals. Advances in computing, data availability, and self-supervised learning (SSL) have paved the way for slide-level foundation models (SLFMs) that can improve prediction tasks in low-data regimes. However, current methods under-utilize shared information between tasks and modalities. To overcome this challenge, we propose ModalTune, a novel fine-tuning framework which introduces the Modal Adapter to integrate new modalities without modifying SLFM weights. Additionally, we use large-language models (LLMs) to encode labels as text, capturing semantic relationships across multiple tasks and cancer types in a single training recipe. ModalTune achieves state-of-the-art (SOTA) results against both uni-modal and multi-modal models across four cancer types, jointly improving survival and cancer subtype prediction while remaining competitive in pan-cancer settings. Additionally, we show ModalTune is generalizable to two out-of-distribution (OOD) datasets. To our knowledge, this is the first unified fine-tuning framework for multi-modal, multi-task, and pan-cancer modeling in digital pathology.
Most existing methods for Magnetic Resonance Imaging (MRI) reconstruction
with deep learning use fully supervised training, which assumes that a high
signal-to-noise ratio (SNR), fully sampled dataset is available for training.
In many circumstances, however, such a dataset is highly impractical or even
technically infeasible to acquire. Recently, a number of self-supervised
methods for MR reconstruction have been proposed, which use sub-sampled data
only. However, the majority of such methods, such as Self-Supervised Learning
via Data Undersampling (SSDU), are susceptible to reconstruction errors arising
from noise in the measured data. In response, we propose Robust SSDU, which
provably recovers clean images from noisy, sub-sampled training data by
simultaneously estimating missing k-space samples and denoising the available
samples. Robust SSDU trains the reconstruction network to map from a further
noisy and sub-sampled version of the data to the original, singly noisy and
sub-sampled data, and applies an additive Noisier2Noise correction term at
inference. We also present a related method, Noiser2Full, that recovers clean
images when noisy, fully sampled data is available for training. Both proposed
methods are applicable to any network architecture, straight-forward to
implement and have similar computational cost to standard training. We evaluate
our methods on the multi-coil fastMRI brain dataset with a novel
denoising-specific architecture and find that it performs competitively with a
benchmark trained on clean, fully sampled data.
 University of TorontoUniversity of New South Wales
University of TorontoUniversity of New South Wales University of AmsterdamHeidelberg UniversityVanderbilt University
University of AmsterdamHeidelberg UniversityVanderbilt University Imperial College LondonUniversity of Zurich
Imperial College LondonUniversity of Zurich GoogleUniversity of Bern
GoogleUniversity of Bern University College London
University College London University of OxfordIT University of CopenhagenThe University of EdinburghUniversity of Edinburgh
University of OxfordIT University of CopenhagenThe University of EdinburghUniversity of Edinburgh McGill University
McGill University NVIDIAVector Institute
NVIDIAVector Institute University of PennsylvaniaSunnybrook Research Institute
University of PennsylvaniaSunnybrook Research Institute Stony Brook University
Stony Brook University King’s College LondonUniversity Health NetworkUniversity of IowaUniversity of GenevaMasaryk University
King’s College LondonUniversity Health NetworkUniversity of IowaUniversity of GenevaMasaryk University University of WarwickGerman Cancer Research Center (DKFZ)University of OttawaINSERMRadboud University Medical CenterUiT The Arctic University of NorwayUniversity Medical Center UtrechtHeidelberg University HospitalMila - Quebec Artificial Intelligence InstituteNational Center for Tumor Diseases (NCT)Leiden University Medical CenterHolon Institute of TechnologyIHU StrasbourgBroad Institute of MIT and HarvardUniversity Medicine EssenNational Institutes of HealthUMC UtrechtOttawa Hospital Research InstituteUniversity of Applied Sciences Western Switzerland (HES-SO)National Cancer InstituteFraunhofer MEVISUniversité de Rennes 1Simula Metropolitan Center for Digital EngineeringINRIA Saclay-Île de FrancePerelman School of Medicine at the University of PennsylvaniaUniversity UtrechtEuropean Molecular Biology Laboratory (EMBL)Case Western Reserve University School of MedicineBioQuantKing
s College LondonCONICET – Universidad de Buenos AiresGoogle Health DeepmindKatholieke Universiteit (KU) Leuven
University of WarwickGerman Cancer Research Center (DKFZ)University of OttawaINSERMRadboud University Medical CenterUiT The Arctic University of NorwayUniversity Medical Center UtrechtHeidelberg University HospitalMila - Quebec Artificial Intelligence InstituteNational Center for Tumor Diseases (NCT)Leiden University Medical CenterHolon Institute of TechnologyIHU StrasbourgBroad Institute of MIT and HarvardUniversity Medicine EssenNational Institutes of HealthUMC UtrechtOttawa Hospital Research InstituteUniversity of Applied Sciences Western Switzerland (HES-SO)National Cancer InstituteFraunhofer MEVISUniversité de Rennes 1Simula Metropolitan Center for Digital EngineeringINRIA Saclay-Île de FrancePerelman School of Medicine at the University of PennsylvaniaUniversity UtrechtEuropean Molecular Biology Laboratory (EMBL)Case Western Reserve University School of MedicineBioQuantKing
s College LondonCONICET – Universidad de Buenos AiresGoogle Health DeepmindKatholieke Universiteit (KU) LeuvenThe "Metrics Reloaded" framework, developed by a large international consortium, provides a systematic, problem-aware guide for selecting and applying performance metrics in biomedical image analysis. It introduces "problem fingerprinting" and a structured decision process to ensure robust and context-specific evaluation of machine learning algorithms.
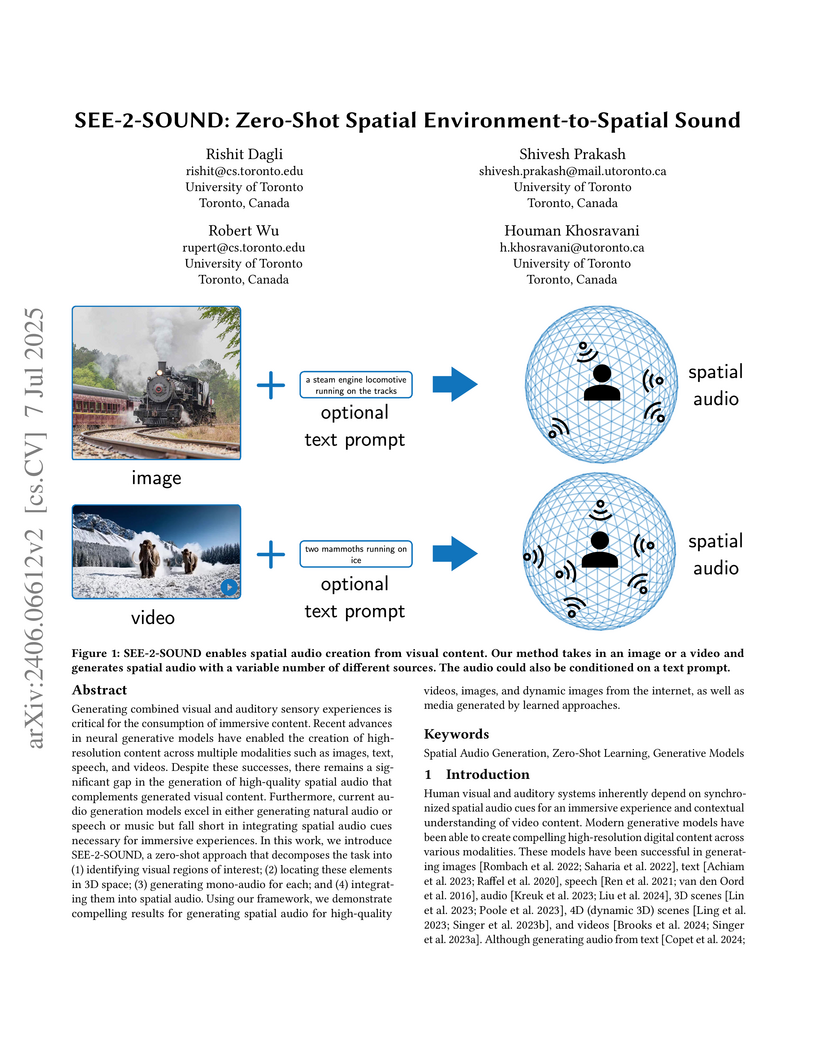
Generating combined visual and auditory sensory experiences is critical for the consumption of immersive content. Recent advances in neural generative models have enabled the creation of high-resolution content across multiple modalities such as images, text, speech, and videos. Despite these successes, there remains a significant gap in the generation of high-quality spatial audio that complements generated visual content. Furthermore, current audio generation models excel in either generating natural audio or speech or music but fall short in integrating spatial audio cues necessary for immersive experiences. In this work, we introduce SEE-2-SOUND, a zero-shot approach that decomposes the task into (1) identifying visual regions of interest; (2) locating these elements in 3D space; (3) generating mono-audio for each; and (4) integrating them into spatial audio. Using our framework, we demonstrate compelling results for generating spatial audio for high-quality videos, images, and dynamic images from the internet, as well as media generated by learned approaches.
 CNRS
CNRS University of Toronto
University of Toronto Imperial College LondonUniversity of Zurich
Imperial College LondonUniversity of Zurich University of Southern CaliforniaUniversity of Bern
University of Southern CaliforniaUniversity of Bern University College London
University College London NVIDIA
NVIDIA KU Leuven
KU Leuven Arizona State UniversitySunnybrook Research Institute
Arizona State UniversitySunnybrook Research Institute King’s College LondonKlinikum rechts der IsarBeijing University of Posts and TelecommunicationsHelmholtz MunichINSA-LyonINSERMTechnical University MunichUniversity of GironaRadboud University Medical CenterPohang University of Science and Technology (POSTECH)Feng Chia UniversityUniversity Medical Center Hamburg-EppendorfUniversity Hospital of ZurichUniversity Hospital BernHelmholtz AICREATISicometrixDeepwise AI LabInselspitalKeck School of MedicineCentre for Medical Image ComputingNational Heart and Lung InstituteWellcome Centre for Human NeuroimagingUniversity Rovira I VirgiliUniversity Institute of Diagnostic and Interventional NeuroradiologyARTORG Center for Biomedical ResearchTranslaTUMStevens Neuroimaging and Informatics InstituteSupport Center of Advanced Neuroimaging (SCAN)Institute for Health SciencesSTADIUS Center for Dynamical Systems, Signal Processing, and Data AnalyticsGraduate School of Artificial Intelligence (GSAI)Institute of Computer Vision and RoboticsLaboratory of Neuro ImagingUniversit ´e Lyon1China Medical University Hsinchu HospitalHurvitz Brain Sciences Research ProgramCenter for Translational Cancer Research
King’s College LondonKlinikum rechts der IsarBeijing University of Posts and TelecommunicationsHelmholtz MunichINSA-LyonINSERMTechnical University MunichUniversity of GironaRadboud University Medical CenterPohang University of Science and Technology (POSTECH)Feng Chia UniversityUniversity Medical Center Hamburg-EppendorfUniversity Hospital of ZurichUniversity Hospital BernHelmholtz AICREATISicometrixDeepwise AI LabInselspitalKeck School of MedicineCentre for Medical Image ComputingNational Heart and Lung InstituteWellcome Centre for Human NeuroimagingUniversity Rovira I VirgiliUniversity Institute of Diagnostic and Interventional NeuroradiologyARTORG Center for Biomedical ResearchTranslaTUMStevens Neuroimaging and Informatics InstituteSupport Center of Advanced Neuroimaging (SCAN)Institute for Health SciencesSTADIUS Center for Dynamical Systems, Signal Processing, and Data AnalyticsGraduate School of Artificial Intelligence (GSAI)Institute of Computer Vision and RoboticsLaboratory of Neuro ImagingUniversit ´e Lyon1China Medical University Hsinchu HospitalHurvitz Brain Sciences Research ProgramCenter for Translational Cancer ResearchDiffusion-weighted MRI (DWI) is essential for stroke diagnosis, treatment decisions, and prognosis. However, image and disease variability hinder the development of generalizable AI algorithms with clinical value. We address this gap by presenting a novel ensemble algorithm derived from the 2022 Ischemic Stroke Lesion Segmentation (ISLES) challenge. ISLES'22 provided 400 patient scans with ischemic stroke from various medical centers, facilitating the development of a wide range of cutting-edge segmentation algorithms by the research community. Through collaboration with leading teams, we combined top-performing algorithms into an ensemble model that overcomes the limitations of individual solutions. Our ensemble model achieved superior ischemic lesion detection and segmentation accuracy on our internal test set compared to individual algorithms. This accuracy generalized well across diverse image and disease variables. Furthermore, the model excelled in extracting clinical biomarkers. Notably, in a Turing-like test, neuroradiologists consistently preferred the algorithm's segmentations over manual expert efforts, highlighting increased comprehensiveness and precision. Validation using a real-world external dataset (N=1686) confirmed the model's generalizability. The algorithm's outputs also demonstrated strong correlations with clinical scores (admission NIHSS and 90-day mRS) on par with or exceeding expert-derived results, underlining its clinical relevance. This study offers two key findings. First, we present an ensemble algorithm (this https URL) that detects and segments ischemic stroke lesions on DWI across diverse scenarios on par with expert (neuro)radiologists. Second, we show the potential for biomedical challenge outputs to extend beyond the challenge's initial objectives, demonstrating their real-world clinical applicability.
This study assesses deep learning models for audio classification in a clinical setting with the constraint of small datasets reflecting real-world prospective data collection. We analyze CNNs, including DenseNet and ConvNeXt, alongside transformer models like ViT, SWIN, and AST, and compare them against pre-trained audio models such as YAMNet and VGGish. Our method highlights the benefits of pre-training on large datasets before fine-tuning on specific clinical data. We prospectively collected two first-of-their-kind patient audio datasets from stroke patients. We investigated various preprocessing techniques, finding that RGB and grayscale spectrogram transformations affect model performance differently based on the priors they learn from pre-training. Our findings indicate CNNs can match or exceed transformer models in small dataset contexts, with DenseNet-Contrastive and AST models showing notable performance. This study highlights the significance of incremental marginal gains through model selection, pre-training, and preprocessing in sound classification; this offers valuable insights for clinical diagnostics that rely on audio classification.
Researchers from the University of Toronto and Sunnybrook Research Institute developed ReCoV and fastReCoV, algorithms that detect noisy labels by analyzing performance fluctuations across repeated cross-validation folds. These methods, including the first explicit noise detection for survival analysis, improved model concordance index on medical data and achieved superior accuracy on classification tasks compared to existing techniques.
Purpose: The scarcity of high-quality curated labeled medical training data
remains one of the major limitations in applying artificial intelligence (AI)
systems to breast cancer diagnosis. Deep models for mammogram analysis and mass
(or micro-calcification) detection require training with a large volume of
labeled images, which are often expensive and time-consuming to collect. To
reduce this challenge, we proposed a novel method that leverages
self-supervised learning (SSL) and a deep hybrid model, named \textbf{HybMNet},
which combines local self-attention and fine-grained feature extraction to
enhance breast cancer detection on screening mammograms.
Approach: Our method employs a two-stage learning process: (1) SSL
Pretraining: We utilize EsViT, a SSL technique, to pretrain a Swin Transformer
(Swin-T) using a limited set of mammograms. The pretrained Swin-T then serves
as the backbone for the downstream task. (2) Downstream Training: The proposed
HybMNet combines the Swin-T backbone with a CNN-based network and a novel
fusion strategy. The Swin-T employs local self-attention to identify
informative patch regions from the high-resolution mammogram, while the
CNN-based network extracts fine-grained local features from the selected
patches. A fusion module then integrates global and local information from both
networks to generate robust predictions. The HybMNet is trained end-to-end,
with the loss function combining the outputs of the Swin-T and CNN modules to
optimize feature extraction and classification performance.
Results: The proposed method was evaluated for its ability to detect breast
cancer by distinguishing between benign (normal) and malignant mammograms.
Leveraging SSL pretraining and the HybMNet model, it achieved AUC of 0.864 (95%
CI: 0.852, 0.875) on the CMMD dataset and 0.889 (95% CI: 0.875, 0.903) on the
INbreast dataset, highlighting its effectiveness.
Ultrasound images are widespread in medical diagnosis for musculoskeletal, cardiac, and obstetrical imaging due to the efficiency and non-invasiveness of the acquisition methodology. However, the acquired images are degraded by acoustic (e.g. reverberation and clutter) and electronic sources of noise. To improve the Peak Signal to Noise Ratio (PSNR) of the images, previous denoising methods often remove the speckles, which could be informative for radiologists and also for quantitative ultrasound. Herein, a method based on the recent Denoising Diffusion Probabilistic Models (DDPM) is proposed. It iteratively enhances the image quality by eliminating the noise while preserving the speckle texture. It is worth noting that the proposed method is trained in a completely unsupervised manner, and no annotated data is required. The experimental blind test results show that our method outperforms the previous nonlocal means denoising methods in terms of PSNR and Generalized Contrast to Noise Ratio (GCNR) while preserving speckles.
Study Objectives: We investigate a Mamba-based deep learning approach for sleep staging on signals from ANNE One (Sibel Health, Evanston, IL), a non-intrusive dual-module wireless wearable system measuring chest electrocardiography (ECG), triaxial accelerometry, and chest temperature, and finger photoplethysmography and finger temperature.
Methods: We obtained wearable sensor recordings from 357 adults undergoing concurrent polysomnography (PSG) at a tertiary care sleep lab. Each PSG recording was manually scored and these annotations served as ground truth labels for training and evaluation of our models. PSG and wearable sensor data were automatically aligned using their ECG channels with manual confirmation by visual inspection. We trained a Mamba-based recurrent neural network architecture on these recordings. Ensembling of model variants with similar architectures was performed.
Results: After ensembling, the model attains a 3-class (wake, non rapid eye movement [NREM] sleep, rapid eye movement [REM] sleep) balanced accuracy of 84.02%, F1 score of 84.23%, Cohen's κ of 72.89%, and a Matthews correlation coefficient (MCC) score of 73.00%; a 4-class (wake, light NREM [N1/N2], deep NREM [N3], REM) balanced accuracy of 75.30%, F1 score of 74.10%, Cohen's κ of 61.51%, and MCC score of 61.95%; a 5-class (wake, N1, N2, N3, REM) balanced accuracy of 65.11%, F1 score of 66.15%, Cohen's κ of 53.23%, MCC score of 54.38%.
Conclusions: Our Mamba-based deep learning model can successfully infer major sleep stages from the ANNE One, a wearable system without electroencephalography (EEG), and can be applied to data from adults attending a tertiary care sleep clinic.
Accurate detection of breast cancer from high-resolution mammograms is
crucial for early diagnosis and effective treatment planning. Previous studies
have shown the potential of using single-view mammograms for breast cancer
detection. However, incorporating multi-view data can provide more
comprehensive insights. Multi-view classification, especially in medical
imaging, presents unique challenges, particularly when dealing with
large-scale, high-resolution data. In this work, we propose a novel Multi-view
Visual Prompt Tuning Network (MVPT-NET) for analyzing multiple screening
mammograms. We first pretrain a robust single-view classification model on
high-resolution mammograms and then innovatively adapt multi-view feature
learning into a task-specific prompt tuning process. This technique selectively
tunes a minimal set of trainable parameters (7\%) while retaining the
robustness of the pre-trained single-view model, enabling efficient integration
of multi-view data without the need for aggressive downsampling. Our approach
offers an efficient alternative to traditional feature fusion methods,
providing a more robust, scalable, and efficient solution for high-resolution
mammogram analysis. Experimental results on a large multi-institution dataset
demonstrate that our method outperforms conventional approaches while
maintaining detection efficiency, achieving an AUROC of 0.852 for
distinguishing between Benign, DCIS, and Invasive classes. This work highlights
the potential of MVPT-NET for medical imaging tasks and provides a scalable
solution for integrating multi-view data in breast cancer detection.
Whole slide images (WSIs) pose unique challenges when training deep learning models. They are very large which makes it necessary to break each image down into smaller patches for analysis, image features have to be extracted at multiple scales in order to capture both detail and context, and extreme class imbalances may exist. Significant progress has been made in the analysis of these images, thanks largely due to the availability of public annotated datasets. We postulate, however, that even if a method scores well on a challenge task, this success may not translate to good performance in a more clinically relevant workflow. Many datasets consist of image patches which may suffer from data curation bias; other datasets are only labelled at the whole slide level and the lack of annotations across an image may mask erroneous local predictions so long as the final decision is correct. In this paper, we outline the differences between patch or slide-level classification versus methods that need to localize or segment cancer accurately across the whole slide, and we experimentally verify that best practices differ in both cases. We apply a binary cancer detection network on post neoadjuvant therapy breast cancer WSIs to find the tumor bed outlining the extent of cancer, a task which requires sensitivity and precision across the whole slide. We extensively study multiple design choices and their effects on the outcome, including architectures and augmentations. Furthermore, we propose a negative data sampling strategy, which drastically reduces the false positive rate (7% on slide level) and improves each metric pertinent to our problem, with a 15% reduction in the error of tumor extent.
Recently there has been an increasing trend to use deep learning frameworks
for both 2D consumer images and for 3D medical images. However, there has been
little effort to use deep frameworks for volumetric vascular segmentation. We
wanted to address this by providing a freely available dataset of 12 annotated
two-photon vasculature microscopy stacks. We demonstrated the use of deep
learning framework consisting both 2D and 3D convolutional filters (ConvNet).
Our hybrid 2D-3D architecture produced promising segmentation result. We
derived the architectures from Lee et al. who used the ZNN framework initially
designed for electron microscope image segmentation. We hope that by sharing
our volumetric vasculature datasets, we will inspire other researchers to
experiment with vasculature dataset and improve the used network architectures.
Self-supervised learning (SSL) has recently shown tremendous potential to
learn generic visual representations useful for many image analysis tasks.
Despite their notable success, the existing SSL methods fail to generalize to
downstream tasks when the number of labeled training instances is small or if
the domain shift between the transfer domains is significant. In this paper, we
attempt to improve self-supervised pretrained representations through the lens
of curriculum learning by proposing a hardness-aware dynamic curriculum
learning (HaDCL) approach. To improve the robustness and generalizability of
SSL, we dynamically leverage progressive harder examples via easy-to-hard and
hard-to-very-hard samples during mini-batch downstream fine-tuning. We discover
that by progressive stage-wise curriculum learning, the pretrained
representations are significantly enhanced and adaptable to both in-domain and
out-of-domain distribution data.
We performed extensive validation on three histology benchmark datasets on
both patch-wise and slide-level classification problems. Our curriculum based
fine-tuning yields a significant improvement over standard fine-tuning, with a
minimum improvement in area-under-the-curve (AUC) score of 1.7% and 2.2% on
in-domain and out-of-domain distribution data, respectively. Further, we
empirically show that our approach is more generic and adaptable to any SSL
methods and does not impose any additional overhead complexity. Besides, we
also outline the role of patch-based versus slide-based curriculum learning in
histopathology to provide practical insights into the success of curriculum
based fine-tuning of SSL methods. Code is released at
this https URL
We present SurgeonAssist-Net: a lightweight framework making action-and-workflow-driven virtual assistance, for a set of predefined surgical tasks, accessible to commercially available optical see-through head-mounted displays (OST-HMDs). On a widely used benchmark dataset for laparoscopic surgical workflow, our implementation competes with state-of-the-art approaches in prediction accuracy for automated task recognition, and yet requires 7.4x fewer parameters, 10.2x fewer floating point operations per second (FLOPS), is 7.0x faster for inference on a CPU, and is capable of near real-time performance on the Microsoft HoloLens 2 OST-HMD. To achieve this, we make use of an efficient convolutional neural network (CNN) backbone to extract discriminative features from image data, and a low-parameter recurrent neural network (RNN) architecture to learn long-term temporal dependencies. To demonstrate the feasibility of our approach for inference on the HoloLens 2 we created a sample dataset that included video of several surgical tasks recorded from a user-centric point-of-view. After training, we deployed our model and cataloged its performance in an online simulated surgical scenario for the prediction of the current surgical task. The utility of our approach is explored in the discussion of several relevant clinical use-cases. Our code is publicly available at this https URL.
High predictive performance and ease of use and interpretability are
important requirements for the applicability of a computer-aided diagnosis
(CAD) to human reading studies. We propose a CAD system specifically designed
to be more comprehensible to the radiologist reviewing screening breast MRI
studies. Multiparametric imaging features are combined to produce a CAD system
for differentiating cancerous and non-cancerous lesions. The complete system
uses a rule-extraction algorithm to present lesion classification results in an
easy to understand graph visualization.
As an analytic pipeline for quantitative imaging feature extraction and
analysis, radiomics has grown rapidly in the past a few years. Recent studies
in radiomics aim to investigate the relationship between tumors imaging
features and clinical outcomes. Open source radiomics feature banks enable the
extraction and analysis of thousands of predefined features. On the other hand,
recent advances in deep learning have shown significant potential in the
quantitative medical imaging field, raising the research question of whether
predefined radiomics features have predictive information in addition to deep
learning features. In this study, we propose a feature fusion method and
investigate whether a combined feature bank of deep learning and predefined
radiomics features can improve the prognostics performance. CT images from
resectable Pancreatic Adenocarcinoma (PDAC) patients were used to compare the
prognosis performance of common feature reduction and fusion methods and the
proposed risk-score based feature fusion method for overall survival. It was
shown that the proposed feature fusion method significantly improves the
prognosis performance for overall survival in resectable PDAC cohorts,
elevating the area under ROC curve by 51% compared to predefined radiomics
features alone, by 16% compared to deep learning features alone, and by 32%
compared to existing feature fusion and reduction methods for a combination of
deep learning and predefined radiomics features.
Objective: Convolutional neural networks (CNNs) have demonstrated promise in
automated cardiac magnetic resonance image segmentation. However, when using
CNNs in a large real-world dataset, it is important to quantify segmentation
uncertainty and identify segmentations which could be problematic. In this
work, we performed a systematic study of Bayesian and non-Bayesian methods for
estimating uncertainty in segmentation neural networks.
Methods: We evaluated Bayes by Backprop, Monte Carlo Dropout, Deep Ensembles,
and Stochastic Segmentation Networks in terms of segmentation accuracy,
probability calibration, uncertainty on out-of-distribution images, and
segmentation quality control.
Results: We observed that Deep Ensembles outperformed the other methods
except for images with heavy noise and blurring distortions. We showed that
Bayes by Backprop is more robust to noise distortions while Stochastic
Segmentation Networks are more resistant to blurring distortions. For
segmentation quality control, we showed that segmentation uncertainty is
correlated with segmentation accuracy for all the methods. With the
incorporation of uncertainty estimates, we were able to reduce the percentage
of poor segmentation to 5% by flagging 31--48% of the most uncertain
segmentations for manual review, substantially lower than random review without
using neural network uncertainty (reviewing 75--78% of all images).
Conclusion: This work provides a comprehensive evaluation of uncertainty
estimation methods and showed that Deep Ensembles outperformed other methods in
most cases.
Significance: Neural network uncertainty measures can help identify
potentially inaccurate segmentations and alert users for manual review.
28 Feb 2018
Shoulder Physiotherapy Exercise Recognition: Machine Learning the Inertial Signals from a Smartwatch
Shoulder Physiotherapy Exercise Recognition: Machine Learning the Inertial Signals from a Smartwatch
Objective: Participation in a physical therapy program is considered one of
the greatest predictors of successful conservative management of common
shoulder disorders. However, adherence to these protocols is often poor and
typically worse for unsupervised home exercise programs. Currently, there are
limited tools available for objective measurement of adherence in the home
setting. The goal of this study was to develop and evaluate the potential for
performing home shoulder physiotherapy monitoring using a commercial
smartwatch.
Approach: Twenty healthy adult subjects with no prior shoulder disorders
performed seven exercises from an evidence-based rotator cuff physiotherapy
protocol, while 6-axis inertial sensor data was collected from the active
extremity. Within an activity recognition chain (ARC) framework, four
supervised learning algorithms were trained and optimized to classify the
exercises: k-nearest neighbor (k-NN), random forest (RF), support vector
machine classifier (SVC), and a convolutional recurrent neural network (CRNN).
Algorithm performance was evaluated using 5-fold cross-validation stratified
first temporally and then by subject.
Main Results: Categorical classification accuracy was above 94% for all
algorithms on the temporally stratified cross validation, with the best
performance achieved by the CRNN algorithm (99.4%). The subject stratified
cross validation, which evaluated classifier performance on unseen subjects,
yielded lower accuracies scores again with CRNN performing best (88.9%).
Significance: This proof of concept study demonstrates the technical
feasibility of a smartwatch device and supervised machine learning approach to
more easily monitor and assess the at-home adherence of shoulder physiotherapy
exercise protocols.
There are no more papers matching your filters at the moment.