University Hospital Bonn
The deployment of Large Language Models (LLM) on mobile devices offers
significant potential for medical applications, enhancing privacy, security,
and cost-efficiency by eliminating reliance on cloud-based services and keeping
sensitive health data local. However, the performance and accuracy of on-device
LLMs in real-world medical contexts remain underexplored. In this study, we
benchmark publicly available on-device LLMs using the AMEGA dataset, evaluating
accuracy, computational efficiency, and thermal limitation across various
mobile devices. Our results indicate that compact general-purpose models like
Phi-3 Mini achieve a strong balance between speed and accuracy, while medically
fine-tuned models such as Med42 and Aloe attain the highest accuracy. Notably,
deploying LLMs on older devices remains feasible, with memory constraints
posing a greater challenge than raw processing power. Our study underscores the
potential of on-device LLMs for healthcare while emphasizing the need for more
efficient inference and models tailored to real-world clinical reasoning.
 University of WashingtonGeorge Washington UniversityIndiana University
University of WashingtonGeorge Washington UniversityIndiana University Cornell University
Cornell University University of California, San Diego
University of California, San Diego Northwestern University
Northwestern University University of PennsylvaniaMassachusetts General Hospital
University of PennsylvaniaMassachusetts General Hospital King’s College London
King’s College London Duke UniversityHelmholtz MunichIntelMayo ClinicUniversity Hospital RWTH AachenUniversity of ZürichTechnical University MunichGerman Cancer Research CenterUniversity of California San FranciscoUniversity Hospital BonnDuke University Medical CenterChildren’s National HospitalKing’s College HospitalChildren’s Hospital of PhiladelphiaSUNY Upstate Medical UniversityGuy’s and St. Thomas’ NHS Foundation TrustSage BionetworksCrestview RadiologyMLCommonsResearch Center JuelichFactored AICMH Lahore Medical CollegeCenter for AI and Data ScienceUniversit
Laval
Duke UniversityHelmholtz MunichIntelMayo ClinicUniversity Hospital RWTH AachenUniversity of ZürichTechnical University MunichGerman Cancer Research CenterUniversity of California San FranciscoUniversity Hospital BonnDuke University Medical CenterChildren’s National HospitalKing’s College HospitalChildren’s Hospital of PhiladelphiaSUNY Upstate Medical UniversityGuy’s and St. Thomas’ NHS Foundation TrustSage BionetworksCrestview RadiologyMLCommonsResearch Center JuelichFactored AICMH Lahore Medical CollegeCenter for AI and Data ScienceUniversit
LavalThe 2024 Brain Tumor Segmentation Meningioma Radiotherapy (BraTS-MEN-RT) challenge aimed to advance automated segmentation algorithms using the largest known multi-institutional dataset of 750 radiotherapy planning brain MRIs with expert-annotated target labels for patients with intact or postoperative meningioma that underwent either conventional external beam radiotherapy or stereotactic radiosurgery. Each case included a defaced 3D post-contrast T1-weighted radiotherapy planning MRI in its native acquisition space, accompanied by a single-label "target volume" representing the gross tumor volume (GTV) and any at-risk post-operative site. Target volume annotations adhered to established radiotherapy planning protocols, ensuring consistency across cases and institutions, and were approved by expert neuroradiologists and radiation oncologists. Six participating teams developed, containerized, and evaluated automated segmentation models using this comprehensive dataset. Team rankings were assessed using a modified lesion-wise Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (95HD). The best reported average lesion-wise DSC and 95HD was 0.815 and 26.92 mm, respectively. BraTS-MEN-RT is expected to significantly advance automated radiotherapy planning by enabling precise tumor segmentation and facilitating tailored treatment, ultimately improving patient outcomes. We describe the design and results from the BraTS-MEN-RT challenge.
Obtaining large pre-trained models that can be fine-tuned to new tasks with limited annotated samples has remained an open challenge for medical imaging data. While pre-trained deep networks on ImageNet and vision-language foundation models trained on web-scale data are prevailing approaches, their effectiveness on medical tasks is limited due to the significant domain shift between natural and medical images. To bridge this gap, we introduce LVM-Med, the first family of deep networks trained on large-scale medical datasets. We have collected approximately 1.3 million medical images from 55 publicly available datasets, covering a large number of organs and modalities such as CT, MRI, X-ray, and Ultrasound. We benchmark several state-of-the-art self-supervised algorithms on this dataset and propose a novel self-supervised contrastive learning algorithm using a graph-matching formulation. The proposed approach makes three contributions: (i) it integrates prior pair-wise image similarity metrics based on local and global information; (ii) it captures the structural constraints of feature embeddings through a loss function constructed via a combinatorial graph-matching objective; and (iii) it can be trained efficiently end-to-end using modern gradient-estimation techniques for black-box solvers. We thoroughly evaluate the proposed LVM-Med on 15 downstream medical tasks ranging from segmentation and classification to object detection, and both for the in and out-of-distribution settings. LVM-Med empirically outperforms a number of state-of-the-art supervised, self-supervised, and foundation models. For challenging tasks such as Brain Tumor Classification or Diabetic Retinopathy Grading, LVM-Med improves previous vision-language models trained on 1 billion masks by 6-7% while using only a ResNet-50.
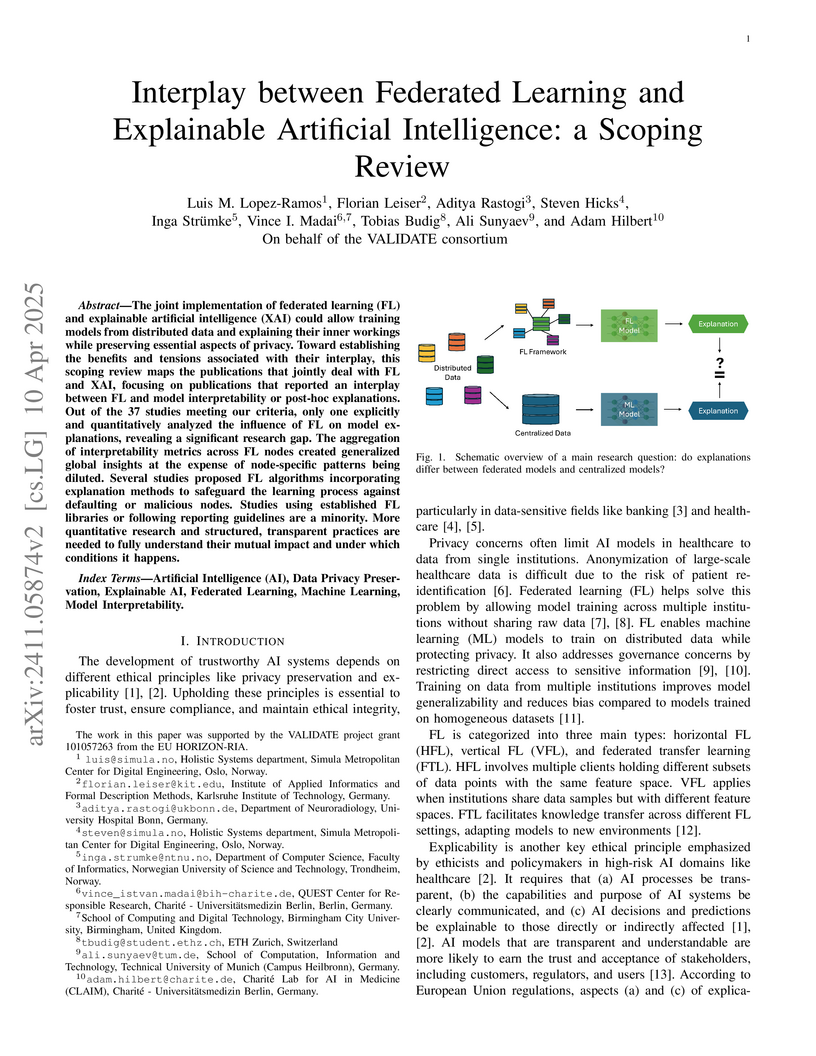
The joint implementation of federated learning (FL) and explainable
artificial intelligence (XAI) could allow training models from distributed data
and explaining their inner workings while preserving essential aspects of
privacy. Toward establishing the benefits and tensions associated with their
interplay, this scoping review maps the publications that jointly deal with FL
and XAI, focusing on publications that reported an interplay between FL and
model interpretability or post-hoc explanations. Out of the 37 studies meeting
our criteria, only one explicitly and quantitatively analyzed the influence of
FL on model explanations, revealing a significant research gap. The aggregation
of interpretability metrics across FL nodes created generalized global insights
at the expense of node-specific patterns being diluted. Several studies
proposed FL algorithms incorporating explanation methods to safeguard the
learning process against defaulting or malicious nodes. Studies using
established FL libraries or following reporting guidelines are a minority. More
quantitative research and structured, transparent practices are needed to fully
understand their mutual impact and under which conditions it happens.
25 Feb 2022
Capturing complex dependence structures between outcome variables (e.g.,
study endpoints) is of high relevance in contemporary biomedical data problems
and medical research. Distributional copula regression provides a flexible tool
to model the joint distribution of multiple outcome variables by disentangling
the marginal response distributions and their dependence structure. In a
regression setup each parameter of the copula model, i.e. the marginal
distribution parameters and the copula dependence parameters, can be related to
covariates via structured additive predictors. We propose a framework to fit
distributional copula regression models via a model-based boosting algorithm.
Model-based boosting is a modern estimation technique that incorporates useful
features like an intrinsic variable selection mechanism, parameter shrinkage
and the capability to fit regression models in high dimensional data setting,
i.e. situations with more covariates than observations. Thus, model-based
boosting does not only complement existing Bayesian and maximum-likelihood
based estimation frameworks for this model class but rather enables unique
intrinsic mechanisms that can be helpful in many applied problems. The
performance of our boosting algorithm in the context of copula regression
models with continuous margins is evaluated in simulation studies that cover
low- and high-dimensional data settings and situations with and without
dependence between the responses. Moreover, distributional copula boosting is
used to jointly analyze and predict the length and the weight of newborns
conditional on sonographic measurements of the fetus before delivery together
with other clinical variables.
Isolated rapid eye movement sleep behavior disorder (iRBD) is a major prodromal marker of α-synucleinopathies, often preceding the clinical onset of Parkinson's disease, dementia with Lewy bodies, or multiple system atrophy. While wrist-worn actimeters hold significant potential for detecting RBD in large-scale screening efforts by capturing abnormal nocturnal movements, they become inoperable without a reliable and efficient analysis pipeline. This study presents ActiTect, a fully automated, open-source machine learning tool to identify RBD from actigraphy recordings. To ensure generalizability across heterogeneous acquisition settings, our pipeline includes robust preprocessing and automated sleep-wake detection to harmonize multi-device data and extract physiologically interpretable motion features characterizing activity patterns. Model development was conducted on a cohort of 78 individuals, yielding strong discrimination under nested cross-validation (AUROC = 0.95). Generalization was confirmed on a blinded local test set (n = 31, AUROC = 0.86) and on two independent external cohorts (n = 113, AUROC = 0.84; n = 57, AUROC = 0.94). To assess real-world robustness, leave-one-dataset-out cross-validation across the internal and external cohorts demonstrated consistent performance (AUROC range = 0.84-0.89). A complementary stability analysis showed that key predictive features remained reproducible across datasets, supporting the final pooled multi-center model as a robust pre-trained resource for broader deployment. By being open-source and easy to use, our tool promotes widespread adoption and facilitates independent validation and collaborative improvements, thereby advancing the field toward a unified and generalizable RBD detection model using wearable devices.
Whole-brain tractography in diffusion MRI is often followed by a parcellation
in which each streamline is classified as belonging to a specific white matter
bundle, or discarded as a false positive. Efficient parcellation is important
both in large-scale studies, which have to process huge amounts of data, and in
the clinic, where computational resources are often limited. TractCloud is a
state-of-the-art approach that aims to maximize accuracy with a local-global
representation. We demonstrate that the local context does not contribute to
the accuracy of that approach, and is even detrimental when dealing with
pathological cases. Based on this observation, we propose PETParc, a new method
for Parallel Efficient Tractography Parcellation. PETParc is a
transformer-based architecture in which the whole-brain tractogram is randomly
partitioned into sub-tractograms whose streamlines are classified in parallel,
while serving as global context for each other. This leads to a speedup of up
to two orders of magnitude relative to TractCloud, and permits inference even
on clinical workstations without a GPU. PETParc accounts for the lack of
streamline orientation either via a novel flip-invariant embedding, or by
simply using flips as part of data augmentation. Despite the speedup, results
are often even better than those of prior methods. The code and pretrained
model will be made public upon acceptance.
Individuals with suspected rare genetic disorders often undergo multiple clinical evaluations, imaging studies, laboratory tests and genetic tests, to find a possible answer over a prolonged period of time. Addressing this "diagnostic odyssey" thus has substantial clinical, psychosocial, and economic benefits. Many rare genetic diseases have distinctive facial features, which can be used by artificial intelligence algorithms to facilitate clinical diagnosis, in prioritizing candidate diseases to be further examined by lab tests or genetic assays, or in helping the phenotype-driven reinterpretation of genome/exome sequencing data. Existing methods using frontal facial photos were built on conventional Convolutional Neural Networks (CNNs), rely exclusively on facial images, and cannot capture non-facial phenotypic traits and demographic information essential for guiding accurate diagnoses. Here we introduce GestaltMML, a multimodal machine learning (MML) approach solely based on the Transformer architecture. It integrates facial images, demographic information (age, sex, ethnicity), and clinical notes (optionally, a list of Human Phenotype Ontology terms) to improve prediction accuracy. Furthermore, we also evaluated GestaltMML on a diverse range of datasets, including 528 diseases from the GestaltMatcher Database, several in-house datasets of Beckwith-Wiedemann syndrome (BWS, over-growth syndrome with distinct facial features), Sotos syndrome (overgrowth syndrome with overlapping features with BWS), NAA10-related neurodevelopmental syndrome, Cornelia de Lange syndrome (multiple malformation syndrome), and KBG syndrome (multiple malformation syndrome). Our results suggest that GestaltMML effectively incorporates multiple modalities of data, greatly narrowing candidate genetic diagnoses of rare diseases and may facilitate the reinterpretation of genome/exome sequencing data.
Portable ultra-low-field MRI (uLF-MRI, 0.064 T) offers accessible neuroimaging for neonatal care but suffers from low signal-to-noise ratio and poor diagnostic quality compared to high-field (HF) MRI. We propose MRIQT, a 3D conditional diffusion framework for image quality transfer (IQT) from uLF to HF MRI. MRIQT combines realistic K-space degradation for physics-consistent uLF simulation, v-prediction with classifier-free guidance for stable image-to-image generation, and an SNR-weighted 3D perceptual loss for anatomical fidelity. The model denoises from a noised uLF input conditioned on the same scan, leveraging volumetric attention-UNet architecture for structure-preserving translation. Trained on a neonatal cohort with diverse pathologies, MRIQT surpasses recent GAN and CNN baselines in PSNR 15.3% with 1.78% over the state of the art, while physicians rated 85% of its outputs as good quality with clear pathology present. MRIQT enables high-fidelity, diffusion-based enhancement of portable ultra-low-field (uLF) MRI for deliable neonatal brain assessment.
Recent advancements in digital pathology have enabled comprehensive analysis of Whole-Slide Images (WSI) from tissue samples, leveraging high-resolution microscopy and computational capabilities. Despite this progress, there is a lack of labeled datasets and open source pipelines specifically tailored for analysis of skin tissue. Here we propose Histo-Miner, a deep learning-based pipeline for analysis of skin WSIs and generate two datasets with labeled nuclei and tumor regions. We develop our pipeline for the analysis of patient samples of cutaneous squamous cell carcinoma (cSCC), a frequent non-melanoma skin cancer. Utilizing the two datasets, comprising 47,392 annotated cell nuclei and 144 tumor-segmented WSIs respectively, both from cSCC patients, Histo-Miner employs convolutional neural networks and vision transformers for nucleus segmentation and classification as well as tumor region segmentation. Performance of trained models positively compares to state of the art with multi-class Panoptic Quality (mPQ) of 0.569 for nucleus segmentation, macro-averaged F1 of 0.832 for nucleus classification and mean Intersection over Union (mIoU) of 0.884 for tumor region segmentation. From these predictions we generate a compact feature vector summarizing tissue morphology and cellular interactions, which can be used for various downstream tasks. Here, we use Histo-Miner to predict cSCC patient response to immunotherapy based on pre-treatment WSIs from 45 patients. Histo-Miner identifies percentages of lymphocytes, the granulocyte to lymphocyte ratio in tumor vicinity and the distances between granulocytes and plasma cells in tumors as predictive features for therapy response. This highlights the applicability of Histo-Miner to clinically relevant scenarios, providing direct interpretation of the classification and insights into the underlying biology.
Over the past decades, computer-aided diagnosis tools for breast cancer have been developed to enhance screening procedures, yet their clinical adoption remains challenged by data variability and inherent biases. Although foundation models (FMs) have recently demonstrated impressive generalizability and transfer learning capabilities by leveraging vast and diverse datasets, their performance can be undermined by spurious correlations that arise from variations in image quality, labeling uncertainty, and sensitive patient attributes. In this work, we explore the fairness and bias of FMs for breast mammography classification by leveraging a large pool of datasets from diverse sources-including data from underrepresented regions and an in-house dataset. Our extensive experiments show that while modality-specific pre-training of FMs enhances performance, classifiers trained on features from individual datasets fail to generalize across domains. Aggregating datasets improves overall performance, yet does not fully mitigate biases, leading to significant disparities across under-represented subgroups such as extreme breast densities and age groups. Furthermore, while domain-adaptation strategies can reduce these disparities, they often incur a performance trade-off. In contrast, fairness-aware techniques yield more stable and equitable performance across subgroups. These findings underscore the necessity of incorporating rigorous fairness evaluations and mitigation strategies into FM-based models to foster inclusive and generalizable AI.
21 Dec 2021
Longitudinal biomedical data are often characterized by a sparse time grid and individual-specific development patterns. Specifically, in epidemiological cohort studies and clinical registries we are facing the question of what can be learned from the data in an early phase of the study, when only a baseline characterization and one follow-up measurement are available. Inspired by recent advances that allow to combine deep learning with dynamic modeling, we investigate whether such approaches can be useful for uncovering complex structure, in particular for an extreme small data setting with only two observations time points for each individual. Irregular spacing in time could then be used to gain more information on individual dynamics by leveraging similarity of individuals. We provide a brief overview of how variational autoencoders (VAEs), as a deep learning approach, can be linked to ordinary differential equations (ODEs) for dynamic modeling, and then specifically investigate the feasibility of such an approach that infers individual-specific latent trajectories by including regularity assumptions and individuals' similarity. We also provide a description of this deep learning approach as a filtering task to give a statistical perspective. Using simulated data, we show to what extent the approach can recover individual trajectories from ODE systems with two and four unknown parameters and infer groups of individuals with similar trajectories, and where it breaks down. The results show that such dynamic deep learning approaches can be useful even in extreme small data settings, but need to be carefully adapted.
Facial analysis technologies have recently measured up to the capabilities of
expert clinicians in syndrome identification. To date, these technologies could
only identify phenotypes of a few diseases, limiting their role in clinical
settings where hundreds of diagnoses must be considered.
We developed a facial analysis framework, DeepGestalt, using computer vision
and deep learning algorithms, that quantifies similarities to hundreds of
genetic syndromes based on unconstrained 2D images. DeepGestalt is currently
trained with over 26,000 patient cases from a rapidly growing
phenotype-genotype database, consisting of tens of thousands of validated
clinical cases, curated through a community-driven platform. DeepGestalt
currently achieves 91% top-10-accuracy in identifying over 215 different
genetic syndromes and has outperformed clinical experts in three separate
experiments.
We suggest that this form of artificial intelligence is ready to support
medical genetics in clinical and laboratory practices and will play a key role
in the future of precision medicine.
19 Apr 2021
A stochastic search method, the so-called Adaptive Subspace (AdaSub) method,
is proposed for variable selection in high-dimensional linear regression
models. The method aims at finding the best model with respect to a certain
model selection criterion and is based on the idea of adaptively solving
low-dimensional sub-problems in order to provide a solution to the original
high-dimensional problem. Any of the usual ℓ0-type model selection
criteria can be used, such as Akaike's Information Criterion (AIC), the
Bayesian Information Criterion (BIC) or the Extended BIC (EBIC), with the last
being particularly suitable for high-dimensional cases. The limiting properties
of the new algorithm are analysed and it is shown that, under certain
conditions, AdaSub converges to the best model according to the considered
criterion. In a simulation study, the performance of AdaSub is investigated in
comparison to alternative methods. The effectiveness of the proposed method is
illustrated via various simulated datasets and a high-dimensional real data
example.
Reconstructing cardiac electrical activity from body surface electric potential measurements results in the severely ill-posed inverse problem in electrocardiography. Many different regularization approaches have been proposed to improve numerical results and provide unique results. This work presents a novel approach for reconstructing the epicardial potential from body surface potential maps based on a space-time total variation-type regularization using finite elements, where a first-order primal-dual algorithm solves the underlying convex optimization problem. In several numerical experiments, the superior performance of this method and the benefit of space-time regularization for the reconstruction of epicardial potential on two-dimensional torso data and a three-dimensional rabbit heart compared to state-of-the-art methods are demonstrated.
Weakly supervised segmentation has the potential to greatly reduce the
annotation effort for training segmentation models for small structures such as
hyper-reflective foci (HRF) in optical coherence tomography (OCT). However,
most weakly supervised methods either involve a strong downsampling of input
images, or only achieve localization at a coarse resolution, both of which are
unsatisfactory for small structures. We propose a novel framework that
increases the spatial resolution of a traditional attention-based Multiple
Instance Learning (MIL) approach by using Layer-wise Relevance Propagation
(LRP) to prompt the Segment Anything Model (SAM~2), and increases recall with
iterative inference. Moreover, we demonstrate that replacing MIL with a Compact
Convolutional Transformer (CCT), which adds a positional encoding, and permits
an exchange of information between different regions of the OCT image, leads to
a further and substantial increase in segmentation accuracy.
Filter-decomposition-based group equivariant convolutional neural networks
(CNNs) have shown promising stability and data efficiency for 3D image feature
extraction. However, these networks, which rely on parameter sharing and
discrete transformation groups, often underperform in modern deep neural
network architectures for processing volumetric images, such as the common 3D
medical images. To address these limitations, this paper presents an efficient
non-parameter-sharing continuous 3D affine group equivariant neural network for
volumetric images. This network uses an adaptive aggregation of Monte Carlo
augmented spherical Fourier-Bessel filter bases to improve the efficiency and
flexibility of 3D group equivariant CNNs for volumetric data. Unlike existing
methods that focus only on angular orthogonality in filter bases, the
introduced spherical Bessel Fourier filter base incorporates both angular and
radial orthogonality to improve feature extraction. Experiments on four medical
image segmentation datasets show that the proposed methods achieve better
affine group equivariance and superior segmentation accuracy than existing 3D
group equivariant convolutional neural network layers, significantly improving
the training stability and data efficiency of conventional CNN layers (at 0.05
significance level). The code is available at
this https URL
19 Nov 2022
Extreme events in a complex network: interplay between degree distribution and repulsive interaction
Extreme events in a complex network: interplay between degree distribution and repulsive interaction
The role of topological heterogeneity in the origin of extreme events in a network is investigated here. The dynamics of the oscillators associated with the nodes are assumed to be identical and influenced by mean-field repulsive interactions. An interplay of topological heterogeneity and the repulsive interaction between the dynamical units of the network triggers extreme events in the nodes when each node succumbs to such events for discretely different ranges of repulsive coupling. A high degree node is vulnerable to weaker repulsive interactions, while a low degree node is susceptible to stronger interactions. As a result, the formation of extreme events changes position with increasing strength of repulsive interaction from high to low degree nodes. Extreme events at any node are identified with the appearance of occasional large-amplitude events (amplitude of the temporal dynamics) that are larger than a threshold height and rare in occurrence, which we confirm by estimating the probability distribution of all events. Extreme events appear at any oscillator near the boundary of transition from rotation to libration at a critical value of the repulsive coupling strength. To explore the phenomenon, a paradigmatic second-order phase model is used to represent the dynamics of the oscillator associated with each node. We make an annealed network approximation to reduce our original model and thereby confirm the dual role of the repulsive interaction and the degree of a node in the origin of extreme events in any oscillator.
 University of TorontoCleveland ClinicUppsala UniversityUniversity Health NetworkTampere UniversityKarolinska Institutet
University of TorontoCleveland ClinicUppsala UniversityUniversity Health NetworkTampere UniversityKarolinska Institutet University of Sydney
University of Sydney Queen Mary University of LondonUniversity of QueenslandUniversity of OtagoUT Southwestern Medical CenterUniversity Hospital BonnYale University School of MedicineAichi Medical UniversityMedical College of WisconsinIndiana University School of MedicineTaipei Veterans General HospitalUniversity Hospital of WalesUniversity of São Paulo Medical SchoolS:t Göran HospitalBostwick LaboratoriesSouthmead HospitalJikei University School of MedicineAquesta Uropathology
Queen Mary University of LondonUniversity of QueenslandUniversity of OtagoUT Southwestern Medical CenterUniversity Hospital BonnYale University School of MedicineAichi Medical UniversityMedical College of WisconsinIndiana University School of MedicineTaipei Veterans General HospitalUniversity Hospital of WalesUniversity of São Paulo Medical SchoolS:t Göran HospitalBostwick LaboratoriesSouthmead HospitalJikei University School of MedicineAquesta UropathologyBackground: An increasing volume of prostate biopsies and a world-wide
shortage of uro-pathologists puts a strain on pathology departments.
Additionally, the high intra- and inter-observer variability in grading can
result in over- and undertreatment of prostate cancer. Artificial intelligence
(AI) methods may alleviate these problems by assisting pathologists to reduce
workload and harmonize grading.
Methods: We digitized 6,682 needle biopsies from 976 participants in the
population based STHLM3 diagnostic study to train deep neural networks for
assessing prostate biopsies. The networks were evaluated by predicting the
presence, extent, and Gleason grade of malignant tissue for an independent test
set comprising 1,631 biopsies from 245 men. We additionally evaluated grading
performance on 87 biopsies individually graded by 23 experienced urological
pathologists from the International Society of Urological Pathology. We
assessed discriminatory performance by receiver operating characteristics (ROC)
and tumor extent predictions by correlating predicted millimeter cancer length
against measurements by the reporting pathologist. We quantified the
concordance between grades assigned by the AI and the expert urological
pathologists using Cohen's kappa.
Results: The performance of the AI to detect and grade cancer in prostate
needle biopsy samples was comparable to that of international experts in
prostate pathology. The AI achieved an area under the ROC curve of 0.997 for
distinguishing between benign and malignant biopsy cores, and 0.999 for
distinguishing between men with or without prostate cancer. The correlation
between millimeter cancer predicted by the AI and assigned by the reporting
pathologist was 0.96. For assigning Gleason grades, the AI achieved an average
pairwise kappa of 0.62. This was within the range of the corresponding values
for the expert pathologists (0.60 to 0.73).
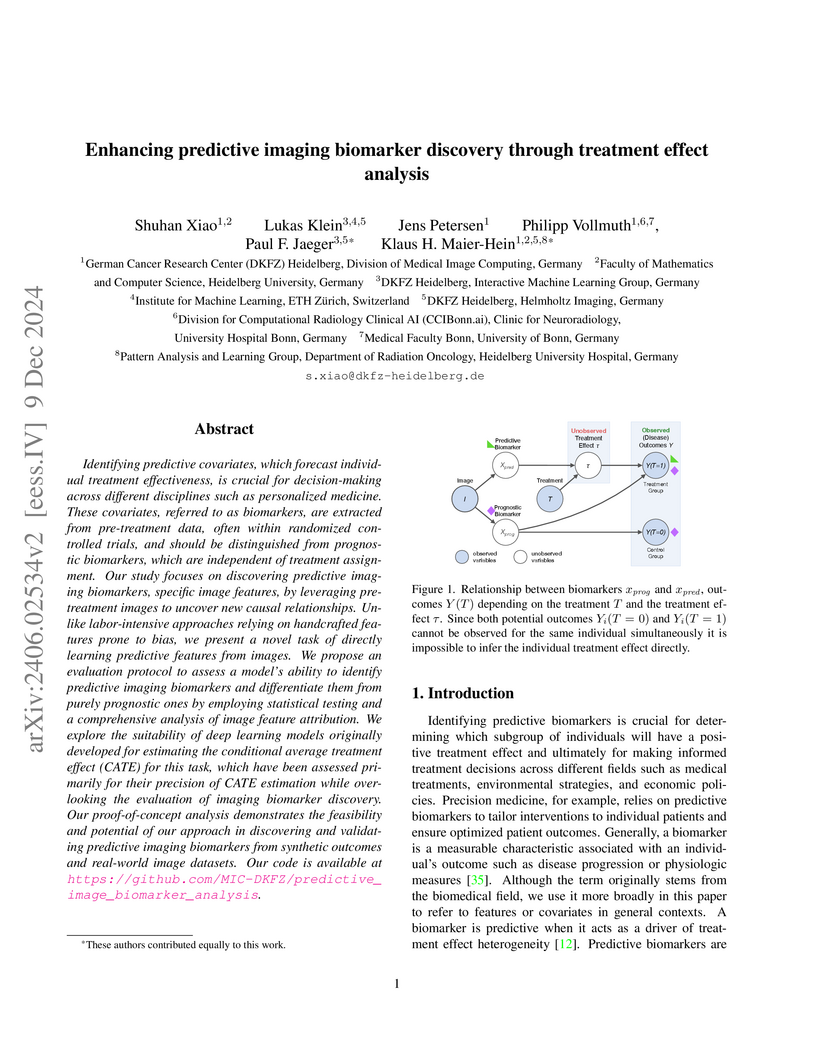
Identifying predictive covariates, which forecast individual treatment effectiveness, is crucial for decision-making across different disciplines such as personalized medicine. These covariates, referred to as biomarkers, are extracted from pre-treatment data, often within randomized controlled trials, and should be distinguished from prognostic biomarkers, which are independent of treatment assignment. Our study focuses on discovering predictive imaging biomarkers, specific image features, by leveraging pre-treatment images to uncover new causal relationships. Unlike labor-intensive approaches relying on handcrafted features prone to bias, we present a novel task of directly learning predictive features from images. We propose an evaluation protocol to assess a model's ability to identify predictive imaging biomarkers and differentiate them from purely prognostic ones by employing statistical testing and a comprehensive analysis of image feature attribution. We explore the suitability of deep learning models originally developed for estimating the conditional average treatment effect (CATE) for this task, which have been assessed primarily for their precision of CATE estimation while overlooking the evaluation of imaging biomarker discovery. Our proof-of-concept analysis demonstrates the feasibility and potential of our approach in discovering and validating predictive imaging biomarkers from synthetic outcomes and real-world image datasets. Our code is available at \url{this https URL}.
There are no more papers matching your filters at the moment.