Cincinnati Children’s Hospital Medical Center
 University of ManchesterUniversity of Zurich
University of ManchesterUniversity of Zurich University of Southern California
University of Southern California McGill University
McGill University Yale University
Yale University University of PennsylvaniaMassachusetts General Hospital
University of PennsylvaniaMassachusetts General Hospital Rutgers UniversityHelmholtz MunichAll India Institute of Medical SciencesBooz Allen HamiltonMayo ClinicTechnical University of DenmarkTechnical University MunichDuke University Medical CenterNational Institutes of HealthChildren’s National HospitalCincinnati Children’s Hospital Medical CenterNationwide Children’s HospitalChildren’s Hospital of PhiladelphiaNational Cancer InstituteUS Food and Drug AdministrationSage BionetworksCrestview RadiologyMercy Catholic Medical CenterChildren’s Health Orange CountyThomas Jefferson University HospitalRoyal Manchester Children’s Hospital
Rutgers UniversityHelmholtz MunichAll India Institute of Medical SciencesBooz Allen HamiltonMayo ClinicTechnical University of DenmarkTechnical University MunichDuke University Medical CenterNational Institutes of HealthChildren’s National HospitalCincinnati Children’s Hospital Medical CenterNationwide Children’s HospitalChildren’s Hospital of PhiladelphiaNational Cancer InstituteUS Food and Drug AdministrationSage BionetworksCrestview RadiologyMercy Catholic Medical CenterChildren’s Health Orange CountyThomas Jefferson University HospitalRoyal Manchester Children’s HospitalPediatric tumors of the central nervous system are the most common cause of cancer-related death in children. The five-year survival rate for high-grade gliomas in children is less than 20%. Due to their rarity, the diagnosis of these entities is often delayed, their treatment is mainly based on historic treatment concepts, and clinical trials require multi-institutional collaborations. Here we present the CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs challenge, focused on pediatric brain tumors with data acquired across multiple international consortia dedicated to pediatric neuro-oncology and clinical trials. The CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs challenge brings together clinicians and AI/imaging scientists to lead to faster development of automated segmentation techniques that could benefit clinical trials, and ultimately the care of children with brain tumors.
15 Jul 2025
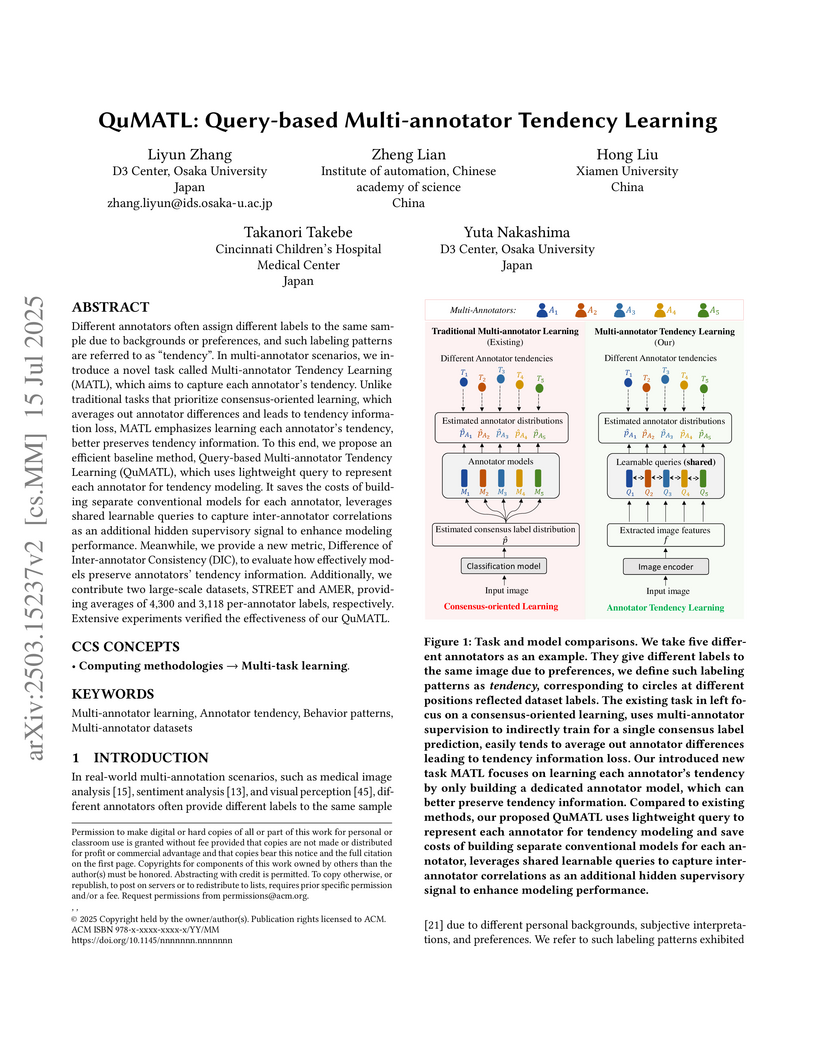
Different annotators often assign different labels to the same sample due to backgrounds or preferences, and such labeling patterns are referred to as tendency. In multi-annotator scenarios, we introduce a novel task called Multi-annotator Tendency Learning (MATL), which aims to capture each annotator tendency. Unlike traditional tasks that prioritize consensus-oriented learning, which averages out annotator differences and leads to tendency information loss, MATL emphasizes learning each annotator tendency, better preserves tendency information. To this end, we propose an efficient baseline method, Query-based Multi-annotator Tendency Learning (QuMATL), which uses lightweight query to represent each annotator for tendency modeling. It saves the costs of building separate conventional models for each annotator, leverages shared learnable queries to capture inter-annotator correlations as an additional hidden supervisory signal to enhance modeling performance. Meanwhile, we provide a new metric, Difference of Inter-annotator Consistency (DIC), to evaluate how effectively models preserve annotators tendency information. Additionally, we contribute two large-scale datasets, STREET and AMER, providing averages of 4300 and 3118 per-annotator labels, respectively. Extensive experiments verified the effectiveness of our QuMATL.
30 Sep 2025
Incorporating historical or real-world data into analyses of treatment effects for rare diseases has become increasingly popular. A major challenge, however, lies in determining the appropriate degree of congruence between historical and current data. In this study, we devote ourselves to the capacity of historical data in replicating the current data, and propose a new congruence measure/estimand pCM. pCM quantifies the heterogeneity between two datasets following the idea of the marginal posterior predictive p-value, and its asymptotic properties were derived. Building upon pCM, we develop the pointwise predictive density calibrated-power prior (PPD-CPP) to dynamically leverage historical information. PPD-CPP achieves the borrowing consistency and allows modeling the power parameter either as a fixed scalar or case-specific quantity informed by covariates. Simulation studies were conducted to demonstrate the performance of these methods and the methodology was illustrated using the Mother's Gift study and \textit{Ceriodaphnia dubia} toxicity test.
Knowledge distillation is a technique to imitate a performance that a deep
learning model has, but reduce the size on another model. It applies the
outputs of a model to train another model having comparable accuracy. These two
distinct models are similar to the way information is delivered in human
society, with one acting as the "teacher" and the other as the "student".
Softmax plays a role in comparing logits generated by models with each other by
converting probability distributions. It delivers the logits of a teacher to a
student with compression through a parameter named temperature. Tuning this
variable reinforces the distillation performance. Although only this parameter
helps with the interaction of logits, it is not clear how temperatures promote
information transfer. In this paper, we propose a novel approach to calculate
the temperature. Our method only refers to the maximum logit generated by a
teacher model, which reduces computational time against state-of-the-art
methods. Our method shows a promising result in different student and teacher
models on a standard benchmark dataset. Algorithms using temperature can obtain
the improvement by plugging in this dynamic approach. Furthermore, the
approximation of the distillation process converges to a correlation of logits
by both models. This reinforces the previous argument that the distillation
conveys the relevance of logits. We report that this approximating algorithm
yields a higher temperature compared to the commonly used static values in
testing.
University of Utah University of ManchesterUniversity of Zurich
University of ManchesterUniversity of Zurich University of Southern California
University of Southern California McGill University
McGill University Yale UniversityHarvard Medical School
Yale UniversityHarvard Medical School University of PennsylvaniaMassachusetts General Hospital
University of PennsylvaniaMassachusetts General Hospital Rutgers UniversityHelmholtz MunichAll India Institute of Medical SciencesBooz Allen HamiltonMayo ClinicTechnical University of DenmarkTechnical University MunichDuke University Medical CenterUniversity of San DiegoBoston Children’s HospitalNational Institutes of HealthChildren’s National HospitalCincinnati Children’s Hospital Medical CenterNational Cancer InstituteIndiana University School of MedicineUS Food and Drug AdministrationSage BionetworksCrestview RadiologyMercy Catholic Medical CenterThe University of Texas Health at HoustonChildren’s Health Orange CountyDana-Farber Brigham Cancer CenterRoyal Manchester Children’s Hospital, Manchester University Hospitals NHS Foundation TrustThomas Jefferson University HospitalThe Children’s Hospital of PhiladelphiaBeth-Israel Deaconess Medical Center
Rutgers UniversityHelmholtz MunichAll India Institute of Medical SciencesBooz Allen HamiltonMayo ClinicTechnical University of DenmarkTechnical University MunichDuke University Medical CenterUniversity of San DiegoBoston Children’s HospitalNational Institutes of HealthChildren’s National HospitalCincinnati Children’s Hospital Medical CenterNational Cancer InstituteIndiana University School of MedicineUS Food and Drug AdministrationSage BionetworksCrestview RadiologyMercy Catholic Medical CenterThe University of Texas Health at HoustonChildren’s Health Orange CountyDana-Farber Brigham Cancer CenterRoyal Manchester Children’s Hospital, Manchester University Hospitals NHS Foundation TrustThomas Jefferson University HospitalThe Children’s Hospital of PhiladelphiaBeth-Israel Deaconess Medical Center
 University of ManchesterUniversity of Zurich
University of ManchesterUniversity of Zurich University of Southern California
University of Southern California McGill University
McGill University Yale UniversityHarvard Medical School
Yale UniversityHarvard Medical School University of PennsylvaniaMassachusetts General Hospital
University of PennsylvaniaMassachusetts General Hospital Rutgers UniversityHelmholtz MunichAll India Institute of Medical SciencesBooz Allen HamiltonMayo ClinicTechnical University of DenmarkTechnical University MunichDuke University Medical CenterUniversity of San DiegoBoston Children’s HospitalNational Institutes of HealthChildren’s National HospitalCincinnati Children’s Hospital Medical CenterNational Cancer InstituteIndiana University School of MedicineUS Food and Drug AdministrationSage BionetworksCrestview RadiologyMercy Catholic Medical CenterThe University of Texas Health at HoustonChildren’s Health Orange CountyDana-Farber Brigham Cancer CenterRoyal Manchester Children’s Hospital, Manchester University Hospitals NHS Foundation TrustThomas Jefferson University HospitalThe Children’s Hospital of PhiladelphiaBeth-Israel Deaconess Medical Center
Rutgers UniversityHelmholtz MunichAll India Institute of Medical SciencesBooz Allen HamiltonMayo ClinicTechnical University of DenmarkTechnical University MunichDuke University Medical CenterUniversity of San DiegoBoston Children’s HospitalNational Institutes of HealthChildren’s National HospitalCincinnati Children’s Hospital Medical CenterNational Cancer InstituteIndiana University School of MedicineUS Food and Drug AdministrationSage BionetworksCrestview RadiologyMercy Catholic Medical CenterThe University of Texas Health at HoustonChildren’s Health Orange CountyDana-Farber Brigham Cancer CenterRoyal Manchester Children’s Hospital, Manchester University Hospitals NHS Foundation TrustThomas Jefferson University HospitalThe Children’s Hospital of PhiladelphiaBeth-Israel Deaconess Medical CenterPediatric central nervous system tumors are the leading cause of cancer-related deaths in children. The five-year survival rate for high-grade glioma in children is less than 20%. The development of new treatments is dependent upon multi-institutional collaborative clinical trials requiring reproducible and accurate centralized response assessment. We present the results of the BraTS-PEDs 2023 challenge, the first Brain Tumor Segmentation (BraTS) challenge focused on pediatric brain tumors. This challenge utilized data acquired from multiple international consortia dedicated to pediatric neuro-oncology and clinical trials. BraTS-PEDs 2023 aimed to evaluate volumetric segmentation algorithms for pediatric brain gliomas from magnetic resonance imaging using standardized quantitative performance evaluation metrics employed across the BraTS 2023 challenges. The top-performing AI approaches for pediatric tumor analysis included ensembles of nnU-Net and Swin UNETR, Auto3DSeg, or nnU-Net with a self-supervised framework. The BraTSPEDs 2023 challenge fostered collaboration between clinicians (neuro-oncologists, neuroradiologists) and AI/imaging scientists, promoting faster data sharing and the development of automated volumetric analysis techniques. These advancements could significantly benefit clinical trials and improve the care of children with brain tumors.
Introduction: Healthcare AI models often inherit biases from their training
data. While efforts have primarily targeted bias in structured data, mental
health heavily depends on unstructured data. This study aims to detect and
mitigate linguistic differences related to non-biological differences in the
training data of AI models designed to assist in pediatric mental health
screening. Our objectives are: (1) to assess the presence of bias by evaluating
outcome parity across sex subgroups, (2) to identify bias sources through
textual distribution analysis, and (3) to develop a de-biasing method for
mental health text data. Methods: We examined classification parity across
demographic groups and assessed how gendered language influences model
predictions. A data-centric de-biasing method was applied, focusing on
neutralizing biased terms while retaining salient clinical information. This
methodology was tested on a model for automatic anxiety detection in pediatric
patients. Results: Our findings revealed a systematic under-diagnosis of female
adolescent patients, with a 4% lower accuracy and a 9% higher False Negative
Rate (FNR) compared to male patients, likely due to disparities in information
density and linguistic differences in patient notes. Notes for male patients
were on average 500 words longer, and linguistic similarity metrics indicated
distinct word distributions between genders. Implementing our de-biasing
approach reduced diagnostic bias by up to 27%, demonstrating its effectiveness
in enhancing equity across demographic groups. Discussion: We developed a
data-centric de-biasing framework to address gender-based content disparities
within clinical text. By neutralizing biased language and enhancing focus on
clinically essential information, our approach demonstrates an effective
strategy for mitigating bias in AI healthcare models trained on text.
Federated learning (FL) is a system in which a central aggregator coordinates
the efforts of multiple clients to solve machine learning problems. This
setting allows training data to be dispersed in order to protect privacy. The
purpose of this paper is to provide an overview of FL systems with a focus on
healthcare. FL is evaluated here based on its frameworks, architectures, and
applications. It is shown here that FL solves the preceding issues with a
shared global deep learning (DL) model via a central aggregator server. This
paper examines recent developments and provides a comprehensive list of
unresolved issues, inspired by the rapid growth of FL research. In the context
of FL, several privacy methods are described, including secure multiparty
computation, homomorphic encryption, differential privacy, and stochastic
gradient descent. Furthermore, a review of various FL classes, such as
horizontal and vertical FL and federated transfer learning, is provided. FL has
applications in wireless communication, service recommendation, intelligent
medical diagnosis systems, and healthcare, all of which are discussed in this
paper. We also present a thorough review of existing FL challenges, such as
privacy protection, communication cost, system heterogeneity, and unreliable
model upload, followed by future research directions.
The integration of artificial intelligence (AI) with radiology marks a
transformative era in medicine. Vision foundation models have been adopted to
enhance radiologic imaging analysis. However, the distinct complexities of
radiologic 2D and 3D radiologic data pose unique challenges that existing
models, pre-trained on general non-medical images, fail to address adequately.
To bridge this gap and capitalize on the diagnostic precision required in
radiologic imaging, we introduce Radiologic Contrastive Language-Image
Pre-training (RadCLIP): a cross-modal vision-language foundational model that
harnesses Vision Language Pre-training (VLP) framework to improve radiologic
image analysis. Building upon Contrastive Language-Image Pre-training (CLIP),
RadCLIP incorporates a slice pooling mechanism tailored for volumetric image
analysis and is pre-trained using a large and diverse dataset of radiologic
image-text pairs. The RadCLIP was pre-trained to effectively align radiologic
images with their corresponding text annotations, creating a robust vision
backbone for radiologic images. Extensive experiments demonstrate RadCLIP's
superior performance in both uni-modal radiologic image classification and
cross-modal image-text matching, highlighting its significant promise for
improving diagnostic accuracy and efficiency in clinical settings. Our Key
contributions include curating a large dataset with diverse radiologic 2D/3D
radiologic image-text pairs, a slice pooling adapter using an attention
mechanism for integrating 2D images, and comprehensive evaluations of RadCLIP
on various radiologic downstream tasks.
It has been hypothesized that one of the main reasons evolution has been able
to produce such impressive adaptations is because it has improved its own
ability to evolve -- "the evolution of evolvability". Rupert Riedl, for
example, an early pioneer of evolutionary developmental biology, suggested that
the evolution of complex adaptations is facilitated by a developmental
organization that is itself shaped by past selection to facilitate evolutionary
innovation. However, selection for characteristics that enable future
innovation seems paradoxical: natural selection cannot favor structures for
benefits they have not yet produced, and favoring characteristics for benefits
that have already been produced does not constitute future innovation. Here we
resolve this paradox by exploiting a formal equivalence between the evolution
of evolvability and learning systems. We use the conditions that enable simple
learning systems to generalize, i.e., to use past experience to improve
performance on previously unseen, future test cases, to demonstrate conditions
where natural selection can systematically favor developmental organizations
that benefit future evolvability. Using numerical simulations of evolution on
highly epistatic fitness landscapes, we illustrate how the structure of evolved
gene regulation networks can result in increased evolvability capable of
avoiding local fitness peaks and discovering higher fitness phenotypes. Our
findings support Riedl's intuition: Developmental organizations that "mimic"
the organization of constraints on phenotypes can be favored by short-term
selection and also facilitate future innovation. Importantly, the conditions
that enable the evolution of such surprising evolvability follow from the same
non-mysterious conditions that permit generalization in learning systems.
18 Aug 2014
We propose a novel "tree-averaging" model that utilizes the ensemble of classification and regression trees (CART). Each constituent tree is estimated with a subset of similar data. We treat this grouping of subsets as Bayesian ensemble trees (BET) and model them as an infinite mixture Dirichlet process. We show that BET adapts to data heterogeneity and accurately estimates each component. Compared with the bootstrap-aggregating approach, BET shows improved prediction performance with fewer trees. We develop an efficient estimating procedure with improved sampling strategies in both CART and mixture models. We demonstrate these advantages of BET with simulations, classification of breast cancer and regression of lung function measurement of cystic fibrosis patients.
Keywords: Bayesian CART; Dirichlet Process; Ensemble Approach; Heterogeneity; Mixture of Trees.
Artificial intelligence based predictive models trained on the clinical notes can be demographically biased. This could lead to adverse healthcare disparities in predicting outcomes like length of stay of the patients. Thus, it is necessary to mitigate the demographic biases within these models. We proposed an implicit in-processing debiasing method to combat disparate treatment which occurs when the machine learning model predict different outcomes for individuals based on the sensitive attributes like gender, ethnicity, race, and likewise. For this purpose, we used clinical notes of heart failure patients and used diagnostic codes, procedure reports and physiological vitals of the patients. We used Clinical BERT to obtain feature embeddings within the diagnostic codes and procedure reports, and LSTM autoencoders to obtain feature embeddings within the physiological vitals. Then, we trained two separate deep learning contrastive learning frameworks, one for gender and the other for ethnicity to obtain debiased representations within those demographic traits. We called this debiasing framework Debias-CLR. We leveraged clinical phenotypes of the patients identified in the diagnostic codes and procedure reports in the previous study to measure fairness statistically. We found that Debias-CLR was able to reduce the Single-Category Word Embedding Association Test (SC-WEAT) effect size score when debiasing for gender and ethnicity. We further found that to obtain fair representations in the embedding space using Debias-CLR, the accuracy of the predictive models on downstream tasks like predicting length of stay of the patients did not get reduced as compared to using the un-debiased counterparts for training the predictive models. Hence, we conclude that our proposed approach, Debias-CLR is fair and representative in mitigating demographic biases and can reduce health disparities.
15 Mar 2025
Within-individual variability of health indicators measured over time is
becoming commonly used to inform about disease progression. Simple summary
statistics (e.g. the standard deviation for each individual) are often used but
they are not suited to account for time changes. In addition, when these
summary statistics are used as covariates in a regression model for
time-to-event outcomes, the estimates of the hazard ratios are subject to
regression dilution. To overcome these issues, a joint model is built where the
association between the time-to-event outcome and multivariate longitudinal
markers is specified in terms of the within-individual variability of the
latter. A mixed-effect location-scale model is used to analyse the longitudinal
biomarkers, their within-individual variability and their correlation. The time
to event is modelled using a proportional hazard regression model, with a
flexible specification of the baseline hazard, and the information from the
longitudinal biomarkers is shared as a function of the random effects. The
model can be used to quantify within-individual variability for the
longitudinal markers and their association with the time-to-event outcome. We
show through a simulation study the performance of the model in comparison with
the standard joint model with constant variance. The model is applied on a
dataset of adult women from the UK cystic fibrosis registry, to evaluate the
association between lung function, malnutrition and mortality.
15 Sep 2025
Instrumental variables are widely used to adjust for measurement error bias when assessing associations of health outcomes with ME prone independent variables. IV approaches addressing ME in longitudinal models are well established, but few methods exist for functional regression. We develop two methods to adjust for ME bias in scalar on function linear models. We regress a scalar outcome on an ME prone functional variable using a functional IV for model identification and propose two least squares based methods to adjust for ME bias. Our methods alleviate potential computational challenges encountered when applying classical regression calibration methods for bias adjustment in high dimensional settings and adjust for potential serial correlations across time. Simulations demonstrate faster run times, lower bias, and lower AIMSE for the proposed methods when compared to existing approaches. The proposed methods were applied to investigate the association between body mass index and wearable device-based physical activity intensity among community dwelling adults living in the United States.
Traditional depression screening methods, such as the PHQ-9, are particularly challenging for children in pediatric primary care due to practical limitations. AI has the potential to help, but the scarcity of annotated datasets in mental health, combined with the computational costs of training, highlights the need for efficient, zero-shot approaches. In this work, we investigate the feasibility of state-of-the-art LLMs for depressive symptom extraction in pediatric settings (ages 6-24). This approach aims to complement traditional screening and minimize diagnostic errors.
Our findings show that all LLMs are 60% more efficient than word match, with Flan leading in precision (average F1: 0.65, precision: 0.78), excelling in the extraction of more rare symptoms like "sleep problems" (F1: 0.92) and "self-loathing" (F1: 0.8). Phi strikes a balance between precision (0.44) and recall (0.60), performing well in categories like "Feeling depressed" (0.69) and "Weight change" (0.78). Llama 3, with the highest recall (0.90), overgeneralizes symptoms, making it less suitable for this type of analysis. Challenges include the complexity of clinical notes and overgeneralization from PHQ-9 scores. The main challenges faced by LLMs include navigating the complex structure of clinical notes with content from different times in the patient trajectory, as well as misinterpreting elevated PHQ-9 scores.
We finally demonstrate the utility of symptom annotations provided by Flan as features in an ML algorithm, which differentiates depression cases from controls with high precision of 0.78, showing a major performance boost compared to a baseline that does not use these features.
07 Aug 2025
Multi-annotator learning (MAL) aims to model annotator-specific labeling patterns. However, existing methods face a critical challenge: they simply skip updating annotator-specific model parameters when encountering missing labels, i.e., a common scenario in real-world crowdsourced datasets where each annotator labels only small subsets of samples. This leads to inefficient data utilization and overfitting risks. To this end, we propose a novel similarity-weighted semi-supervised learning framework (SimLabel) that leverages inter-annotator similarities to generate weighted soft labels for missing annotations, enabling the utilization of unannotated samples rather than skipping them entirely. We further introduce a confidence-based iterative refinement mechanism that combines maximum probability with entropy-based uncertainty to prioritize predicted high-quality pseudo-labels to impute missing labels, jointly enhancing similarity estimation and model performance over time. For evaluation, we contribute a new multimodal multi-annotator dataset, AMER2, with high and more variable missing rates, reflecting real-world annotation sparsity and enabling evaluation across different sparsity levels.
General-purpose clinical natural language processing (NLP) tools are
increasingly used for the automatic labeling of clinical reports. However,
independent evaluations for specific tasks, such as pediatric chest radiograph
(CXR) report labeling, are limited. This study compares four commercial
clinical NLP systems - Amazon Comprehend Medical (AWS), Google Healthcare NLP
(GC), Azure Clinical NLP (AZ), and SparkNLP (SP) - for entity extraction and
assertion detection in pediatric CXR reports. Additionally, CheXpert and
CheXbert, two dedicated chest radiograph report labelers, were evaluated on the
same task using CheXpert-defined labels. We analyzed 95,008 pediatric CXR
reports from a large academic pediatric hospital. Entities and assertion
statuses (positive, negative, uncertain) from the findings and impression
sections were extracted by the NLP systems, with impression section entities
mapped to 12 disease categories and a No Findings category. CheXpert and
CheXbert extracted the same 13 categories. Outputs were compared using Fleiss
Kappa and accuracy against a consensus pseudo-ground truth. Significant
differences were found in the number of extracted entities and assertion
distributions across NLP systems. SP extracted 49,688 unique entities, GC
16,477, AZ 31,543, and AWS 27,216. Assertion accuracy across models averaged
around 62%, with SP highest (76%) and AWS lowest (50%). CheXpert and CheXbert
achieved 56% accuracy. Considerable variability in performance highlights the
need for careful validation and review before deploying NLP tools for clinical
report labeling.
22 Jul 2025
Integrating diverse data sources offers a comprehensive view of patient health and holds potential for improving clinical decision-making. In Cystic Fibrosis (CF), which is a genetic disorder primarily affecting the lungs, biomarkers that track lung function decline such as FEV1 serve as important predictors for assessing disease progression. Prior research has shown that incorporating social and environmental determinants of health improves prognostic accuracy. To investigate the lung function decline among individuals with CF, we integrate data from the U.S. Cystic Fibrosis Foundation Patient Registry with social and environmental health information. Our analysis focuses on the relationship between lung function and the deprivation index, a composite measure of socioeconomic status.
We used advanced multivariate mixed-effects models, which allow for the joint modelling of multiple longitudinal outcomes with flexible functional forms. This methodology provides an understanding of interrelationships among outcomes, addressing the complexities of dynamic health data. We examine whether this relationship varies with patients' exposure duration to high-deprivation areas, analyzing data across time and within individual US states. Results show a strong relation between lung function and the area under the deprivation index curve across all states. These results underscore the importance of integrating social and environmental determinants of health into clinical models of disease progression. By accounting for broader contextual factors, healthcare providers can gain deeper insights into disease trajectories and design more targeted intervention strategies.
07 Aug 2025
Multi-annotator learning traditionally aggregates diverse annotations to approximate a single ground truth, treating disagreements as noise. However, this paradigm faces fundamental challenges: subjective tasks often lack absolute ground truth, and sparse annotation coverage makes aggregation statistically unreliable. We introduce a paradigm shift from sample-wise aggregation to annotator-wise behavior modeling. By treating annotator disagreements as valuable information rather than noise, modeling annotator-specific behavior patterns can reconstruct unlabeled data to reduce annotation cost, enhance aggregation reliability, and explain annotator decision behavior. To this end, we propose QuMAB (Query-based Multi-Annotator Behavior Pattern Learning), which uses light-weight queries to model individual annotators while capturing inter-annotator correlations as implicit regularization, preventing overfitting to sparse individual data while maintaining individualization and improving generalization, with a visualization of annotator focus regions offering an explainable analysis of behavior understanding. We contribute two large-scale datasets with dense per-annotator labels: STREET (4,300 labels/annotator) and AMER (average 3,118 labels/annotator), the first multimodal multi-annotator dataset. Extensive experiments demonstrate the superiority of our QuMAB in modeling individual annotators' behavior patterns, their utility for consensus prediction, and applicability under sparse annotations.
15 Apr 2013
In an optimal nonbipartite match, a single population is divided into matched
pairs to minimize a total distance within matched pairs. Nonbipartite matching
has been used to strengthen instrumental variables in observational studies of
treatment effects, essentially by forming pairs that are similar in terms of
covariates but very different in the strength of encouragement to accept the
treatment. Optimal nonbipartite matching is typically done using network
optimization techniques that can be quick, running in polynomial time, but
these techniques limit the tools available for matching. Instead, we use
integer programming techniques, thereby obtaining a wealth of new tools not
previously available for nonbipartite matching, including fine and near-fine
balance for several nominal variables, forced near balance on means and optimal
subsetting. We illustrate the methods in our on-going study of outcomes of
late-preterm births in California, that is, births of 34 to 36 weeks of
gestation. Would lengthening the time in the hospital for such births reduce
the frequency of rapid readmissions? A straightforward comparison of babies who
stay for a shorter or longer time would be severely biased, because the
principal reason for a long stay is some serious health problem. We need an
instrument, something inconsequential and haphazard that encourages a shorter
or a longer stay in the hospital. It turns out that babies born at certain
times of day tend to stay overnight once with a shorter length of stay, whereas
babies born at other times of day tend to stay overnight twice with a longer
length of stay, and there is nothing particularly special about a baby who is
born at 11:00 pm.
28 Nov 2025
We introduce EEG Autoclean Vision Language AI (ICVision) a first-of-its-kind system that emulates expert-level EEG ICA component classification through AI-agent vision and natural language reasoning. Unlike conventional classifiers such as ICLabel, which rely on handcrafted features, ICVision directly interprets ICA dashboard visualizations topography, time series, power spectra, and ERP plots, using a multimodal large language model (GPT-4 Vision). This allows the AI to see and explain EEG components the way trained neurologists do, making it the first scientific implementation of AI-agent visual cognition in neurophysiology. ICVision classifies each component into one of six canonical categories (brain, eye, heart, muscle, channel noise, and other noise), returning both a confidence score and a human-like explanation. Evaluated on 3,168 ICA components from 124 EEG datasets, ICVision achieved k = 0.677 agreement with expert consensus, surpassing MNE ICLabel, while also preserving clinically relevant brain signals in ambiguous cases. Over 97% of its outputs were rated as interpretable and actionable by expert reviewers. As a core module of the open-source EEG Autoclean platform, ICVision signals a paradigm shift in scientific AI, where models do not just classify, but see, reason, and communicate. It opens the door to globally scalable, explainable, and reproducible EEG workflows, marking the emergence of AI agents capable of expert-level visual decision-making in brain science and beyond.
There are no more papers matching your filters at the moment.