Guy’s and St. Thomas’ NHS Foundation Trust
To make medical datasets accessible without sharing sensitive patient information, we introduce a novel end-to-end approach for generative de-identification of dynamic medical imaging data. Until now, generative methods have faced constraints in terms of fidelity, spatio-temporal coherence, and the length of generation, failing to capture the complete details of dataset distributions. We present a model designed to produce high-fidelity, long and complete data samples with near-real-time efficiency and explore our approach on a challenging task: generating echocardiogram videos. We develop our generation method based on diffusion models and introduce a protocol for medical video dataset anonymization. As an exemplar, we present EchoNet-Synthetic, a fully synthetic, privacy-compliant echocardiogram dataset with paired ejection fraction labels. As part of our de-identification protocol, we evaluate the quality of the generated dataset and propose to use clinical downstream tasks as a measurement on top of widely used but potentially biased image quality metrics. Experimental outcomes demonstrate that EchoNet-Synthetic achieves comparable dataset fidelity to the actual dataset, effectively supporting the ejection fraction regression task. Code, weights and dataset are available at this https URL.
 University of WashingtonGeorge Washington UniversityIndiana University
University of WashingtonGeorge Washington UniversityIndiana University Cornell University
Cornell University University of California, San Diego
University of California, San Diego Northwestern University
Northwestern University University of PennsylvaniaMassachusetts General Hospital
University of PennsylvaniaMassachusetts General Hospital King’s College London
King’s College London Duke UniversityHelmholtz MunichIntelMayo ClinicUniversity Hospital RWTH AachenUniversity of ZürichTechnical University MunichGerman Cancer Research CenterUniversity of California San FranciscoUniversity Hospital BonnDuke University Medical CenterChildren’s National HospitalKing’s College HospitalChildren’s Hospital of PhiladelphiaSUNY Upstate Medical UniversityGuy’s and St. Thomas’ NHS Foundation TrustSage BionetworksCrestview RadiologyMLCommonsResearch Center JuelichFactored AICMH Lahore Medical CollegeCenter for AI and Data ScienceUniversit
Laval
Duke UniversityHelmholtz MunichIntelMayo ClinicUniversity Hospital RWTH AachenUniversity of ZürichTechnical University MunichGerman Cancer Research CenterUniversity of California San FranciscoUniversity Hospital BonnDuke University Medical CenterChildren’s National HospitalKing’s College HospitalChildren’s Hospital of PhiladelphiaSUNY Upstate Medical UniversityGuy’s and St. Thomas’ NHS Foundation TrustSage BionetworksCrestview RadiologyMLCommonsResearch Center JuelichFactored AICMH Lahore Medical CollegeCenter for AI and Data ScienceUniversit
LavalThe 2024 Brain Tumor Segmentation Meningioma Radiotherapy (BraTS-MEN-RT) challenge aimed to advance automated segmentation algorithms using the largest known multi-institutional dataset of 750 radiotherapy planning brain MRIs with expert-annotated target labels for patients with intact or postoperative meningioma that underwent either conventional external beam radiotherapy or stereotactic radiosurgery. Each case included a defaced 3D post-contrast T1-weighted radiotherapy planning MRI in its native acquisition space, accompanied by a single-label "target volume" representing the gross tumor volume (GTV) and any at-risk post-operative site. Target volume annotations adhered to established radiotherapy planning protocols, ensuring consistency across cases and institutions, and were approved by expert neuroradiologists and radiation oncologists. Six participating teams developed, containerized, and evaluated automated segmentation models using this comprehensive dataset. Team rankings were assessed using a modified lesion-wise Dice Similarity Coefficient (DSC) and 95% Hausdorff Distance (95HD). The best reported average lesion-wise DSC and 95HD was 0.815 and 26.92 mm, respectively. BraTS-MEN-RT is expected to significantly advance automated radiotherapy planning by enabling precise tumor segmentation and facilitating tailored treatment, ultimately improving patient outcomes. We describe the design and results from the BraTS-MEN-RT challenge.
Transesophageal echocardiography (TEE) plays a pivotal role in cardiology for diagnostic and interventional procedures. However, using it effectively requires extensive training due to the intricate nature of image acquisition and interpretation. To enhance the efficiency of novice sonographers and reduce variability in scan acquisitions, we propose a novel ultrasound (US) navigation assistance method based on contrastive learning as goal-conditioned reinforcement learning (GCRL). We augment the previous framework using a novel contrastive patient batching method (CPB) and a data-augmented contrastive loss, both of which we demonstrate are essential to ensure generalization to anatomical variations across patients. The proposed framework enables navigation to both standard diagnostic as well as intricate interventional views with a single model. Our method was developed with a large dataset of 789 patients and obtained an average error of 6.56 mm in position and 9.36 degrees in angle on a testing dataset of 140 patients, which is competitive or superior to models trained on individual views. Furthermore, we quantitatively validate our method's ability to navigate to interventional views such as the Left Atrial Appendage (LAA) view used in LAA closure. Our approach holds promise in providing valuable guidance during transesophageal ultrasound examinations, contributing to the advancement of skill acquisition for cardiac ultrasound practitioners.
Image synthesis is expected to provide value for the translation of machine learning methods into clinical practice. Fundamental problems like model robustness, domain transfer, causal modelling, and operator training become approachable through synthetic data. Especially, heavily operator-dependant modalities like Ultrasound imaging require robust frameworks for image and video generation. So far, video generation has only been possible by providing input data that is as rich as the output data, e.g., image sequence plus conditioning in, video out. However, clinical documentation is usually scarce and only single images are reported and stored, thus retrospective patient-specific analysis or the generation of rich training data becomes impossible with current approaches. In this paper, we extend elucidated diffusion models for video modelling to generate plausible video sequences from single images and arbitrary conditioning with clinical parameters. We explore this idea within the context of echocardiograms by looking into the variation of the Left Ventricle Ejection Fraction, the most essential clinical metric gained from these examinations. We use the publicly available EchoNet-Dynamic dataset for all our experiments. Our image to sequence approach achieves an R2 score of 93%, which is 38 points higher than recently proposed sequence to sequence generation methods. Code and models will be available at: this https URL.
With respect to spatial overlap, CNN-based segmentation of short axis
cardiovascular magnetic resonance (CMR) images has achieved a level of
performance consistent with inter observer variation. However, conventional
training procedures frequently depend on pixel-wise loss functions, limiting
optimisation with respect to extended or global features. As a result, inferred
segmentations can lack spatial coherence, including spurious connected
components or holes. Such results are implausible, violating the anticipated
topology of image segments, which is frequently known a priori. Addressing this
challenge, published work has employed persistent homology, constructing
topological loss functions for the evaluation of image segments against an
explicit prior. Building a richer description of segmentation topology by
considering all possible labels and label pairs, we extend these losses to the
task of multi-class segmentation. These topological priors allow us to resolve
all topological errors in a subset of 150 examples from the ACDC short axis CMR
training data set, without sacrificing overlap performance.
Sickle Cell Disease (SCD) is one of the most common genetic diseases in the world. Splenomegaly (abnormal enlargement of the spleen) is frequent among children with SCD. If left untreated, splenomegaly can be life-threatening. The current workflow to measure spleen size includes palpation, possibly followed by manual length measurement in 2D ultrasound imaging. However, this manual measurement is dependent on operator expertise and is subject to intra- and inter-observer variability. We investigate the use of deep learning to perform automatic estimation of spleen length from ultrasound images. We investigate two types of approach, one segmentation-based and one based on direct length estimation, and compare the results against measurements made by human experts. Our best model (segmentation-based) achieved a percentage length error of 7.42%, which is approaching the level of inter-observer variability (5.47%-6.34%). To the best of our knowledge, this is the first attempt to measure spleen size in a fully automated way from ultrasound images.
24 Sep 2020
The initial detection and identification of suspicious lesions and the
precise delineation of tumour mar-gins are essential for successful tumour
resection, with progression-free survival linked to rates of complete
resection. However, post-surgical positive margin rates remain high for many
cancers and despite numerous advances in intraoperative imaging and diagnostic
technologies, there exists no single modality that can adequately perform both
tumoural detection and delineation. Here, we demonstrate a multimodal computer
vision-based diagnostic system capable of both the gross detection and
identification of suspicious lesions and the precise delineation of disease
margins. We first show that through visual tracking of a spectroscopic probe,
we enable real-time tumour margin delineation both for ex vivo human tumour
biopsies and for an in vivo tumour xenograft mouse model. We then demonstrate
that the combination of Raman spectroscopic diagnoses with protoporphyrin IX
(PPIX) fluorescence imaging enables fluorescence-guided Raman spectroscopic
margin delineation. Our fluorescence-guided Raman spectroscopic system achieves
superior margin delineation accuracy to fluorescence imaging alone,
demonstrating the potential for our system to achieve improved clinical
outcomes for tumour resection surgeries.
Multi-class segmentation of cardiac magnetic resonance (CMR) images seeks a
separation of data into anatomical components with known structure and
configuration. The most popular CNN-based methods are optimised using pixel
wise loss functions, ignorant of the spatially extended features that
characterise anatomy. Therefore, whilst sharing a high spatial overlap with the
ground truth, inferred CNN-based segmentations can lack coherence, including
spurious connected components, holes and voids. Such results are implausible,
violating anticipated anatomical topology. In response, (single-class)
persistent homology-based loss functions have been proposed to capture global
anatomical features. Our work extends these approaches to the task of
multi-class segmentation. Building an enriched topological description of all
class labels and class label pairs, our loss functions make predictable and
statistically significant improvements in segmentation topology using a
CNN-based post-processing framework. We also present (and make available) a
highly efficient implementation based on cubical complexes and parallel
execution, enabling practical application within high resolution 3D data for
the first time. We demonstrate our approach on 2D short axis and 3D whole heart
CMR segmentation, advancing a detailed and faithful analysis of performance on
two publicly available datasets.
Artificial Intelligence (AI) has become commonplace to solve routine everyday
tasks. Because of the exponential growth in medical imaging data volume and
complexity, the workload on radiologists is steadily increasing. We project
that the gap between the number of imaging exams and the number of expert
radiologist readers required to cover this increase will continue to expand,
consequently introducing a demand for AI-based tools that improve the
efficiency with which radiologists can comfortably interpret these exams. AI
has been shown to improve efficiency in medical-image generation, processing,
and interpretation, and a variety of such AI models have been developed across
research labs worldwide. However, very few of these, if any, find their way
into routine clinical use, a discrepancy that reflects the divide between AI
research and successful AI translation. To address the barrier to clinical
deployment, we have formed MONAI Consortium, an open-source community which is
building standards for AI deployment in healthcare institutions, and developing
tools and infrastructure to facilitate their implementation. This report
represents several years of weekly discussions and hands-on problem solving
experience by groups of industry experts and clinicians in the MONAI
Consortium. We identify barriers between AI-model development in research labs
and subsequent clinical deployment and propose solutions. Our report provides
guidance on processes which take an imaging AI model from development to
clinical implementation in a healthcare institution. We discuss various AI
integration points in a clinical Radiology workflow. We also present a taxonomy
of Radiology AI use-cases. Through this report, we intend to educate the
stakeholders in healthcare and AI (AI researchers, radiologists, imaging
informaticists, and regulators) about cross-disciplinary challenges and
possible solutions.
Quantum machine learning with quantum kernels for classification problems is
a growing area of research. Recently, quantum kernel alignment techniques that
parameterise the kernel have been developed, allowing the kernel to be trained
and therefore aligned with a specific dataset. While quantum kernel alignment
is a promising technique, it has been hampered by considerable training costs
because the full kernel matrix must be constructed at every training iteration.
Addressing this challenge, we introduce a novel method that seeks to balance
efficiency and performance. We present a sub-sampling training approach that
uses a subset of the kernel matrix at each training step, thereby reducing the
overall computational cost of the training. In this work, we apply the
sub-sampling method to synthetic datasets and a real-world breast cancer
dataset and demonstrate considerable reductions in the number of circuits
required to train the quantum kernel while maintaining classification accuracy.
Purpose: Subarachnoid haemorrhage is a potentially fatal consequence of intracranial aneurysm rupture, however, it is difficult to predict if aneurysms will rupture. Prophylactic treatment of an intracranial aneurysm also involves risk, hence identifying rupture-prone aneurysms is of substantial clinical importance. This systematic review aims to evaluate the performance of machine learning algorithms for predicting intracranial aneurysm rupture risk.
Methods: MEDLINE, Embase, Cochrane Library and Web of Science were searched until December 2023. Studies incorporating any machine learning algorithm to predict the risk of rupture of an intracranial aneurysm were included. Risk of bias was assessed using the Prediction Model Risk of Bias Assessment Tool (PROBAST). PROSPERO registration: CRD42023452509. Results: Out of 10,307 records screened, 20 studies met the eligibility criteria for this review incorporating a total of 20,286 aneurysm cases. The machine learning models gave a 0.66-0.90 range for performance accuracy. The models were compared to current clinical standards in six studies and gave mixed results. Most studies posed high or unclear risks of bias and concerns for applicability, limiting the inferences that can be drawn from them. There was insufficient homogenous data for a meta-analysis.
Conclusions: Machine learning can be applied to predict the risk of rupture for intracranial aneurysms. However, the evidence does not comprehensively demonstrate superiority to existing practice, limiting its role as a clinical adjunct. Further prospective multicentre studies of recent machine learning tools are needed to prove clinical validation before they are implemented in the clinic.
15 Mar 2025
Within-individual variability of health indicators measured over time is
becoming commonly used to inform about disease progression. Simple summary
statistics (e.g. the standard deviation for each individual) are often used but
they are not suited to account for time changes. In addition, when these
summary statistics are used as covariates in a regression model for
time-to-event outcomes, the estimates of the hazard ratios are subject to
regression dilution. To overcome these issues, a joint model is built where the
association between the time-to-event outcome and multivariate longitudinal
markers is specified in terms of the within-individual variability of the
latter. A mixed-effect location-scale model is used to analyse the longitudinal
biomarkers, their within-individual variability and their correlation. The time
to event is modelled using a proportional hazard regression model, with a
flexible specification of the baseline hazard, and the information from the
longitudinal biomarkers is shared as a function of the random effects. The
model can be used to quantify within-individual variability for the
longitudinal markers and their association with the time-to-event outcome. We
show through a simulation study the performance of the model in comparison with
the standard joint model with constant variance. The model is applied on a
dataset of adult women from the UK cystic fibrosis registry, to evaluate the
association between lung function, malnutrition and mortality.
 University of Manchester
University of Manchester University College London
University College London University of OxfordIBM Research
University of OxfordIBM Research King’s College LondonKarolinska InstitutetLondon School of Hygiene and Tropical MedicineGuy’s and St. Thomas’ NHS Foundation TrustLiverpool School of Tropical MedicineThe Alan Turing Institute for Data Science and Artificial IntelligenceUnited Kingdom Health Security Agency (UKHSA)Liverpool University Hospitals NHS Foundation Trust
King’s College LondonKarolinska InstitutetLondon School of Hygiene and Tropical MedicineGuy’s and St. Thomas’ NHS Foundation TrustLiverpool School of Tropical MedicineThe Alan Turing Institute for Data Science and Artificial IntelligenceUnited Kingdom Health Security Agency (UKHSA)Liverpool University Hospitals NHS Foundation TrustThrough the use of cutting-edge unsupervised classification techniques from statistics and machine learning, we characterise symptom phenotypes among symptomatic SARS-CoV-2 PCR-positive community cases. We first analyse each dataset in isolation and across age bands, before using methods that allow us to compare multiple datasets. While we observe separation due to the total number of symptoms experienced by cases, we also see a separation of symptoms into gastrointestinal, respiratory and other types, and different symptom co-occurrence patterns at the extremes of age. In this way, we are able to demonstrate the deep structure of symptoms of COVID-19 without usual biases due to study design. This is expected to have implications for the identification and management of community SARS-CoV-2 cases and could be further applied to symptom-based management of other diseases and syndromes.
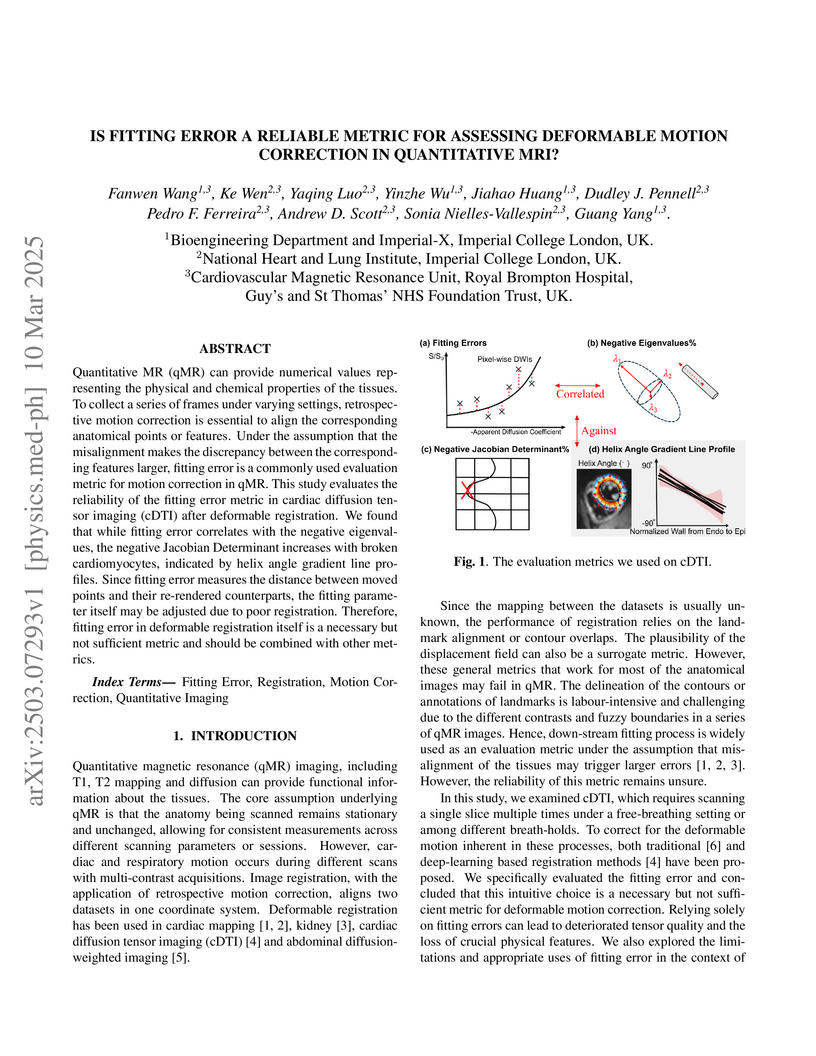
Quantitative MR (qMR) can provide numerical values representing the physical
and chemical properties of the tissues. To collect a series of frames under
varying settings, retrospective motion correction is essential to align the
corresponding anatomical points or features. Under the assumption that the
misalignment makes the discrepancy between the corresponding features larger,
fitting error is a commonly used evaluation metric for motion correction in
qMR. This study evaluates the reliability of the fitting error metric in
cardiac diffusion tensor imaging (cDTI) after deformable registration. We found
that while fitting error correlates with the negative eigenvalues, the negative
Jacobian Determinant increases with broken cardiomyocytes, indicated by helix
angle gradient line profiles. Since fitting error measures the distance between
moved points and their re-rendered counterparts, the fitting parameter itself
may be adjusted due to poor registration. Therefore, fitting error in
deformable registration itself is a necessary but not sufficient metric and
should be combined with other metrics.
23 Jun 2022
Background. Absorbed radiation dose to the mandible is an important risk
factor in the development of mandibular osteoradionecrosis (ORN) in head and
neck cancer (HNC) patients treated with radiotherapy (RT). The prediction of
mandibular ORN may not only guide the RT treatment planning optimisation
process but also identify which patients would benefit from a closer follow-up
post-RT for an early diagnosis and intervention of ORN. Existing mandibular ORN
prediction models are based on dose-volume histogram (DVH) metrics that omit
the spatial localisation and dose gradient and direction information provided
by the clinical mandible radiation dose distribution maps. Methods. We propose
the use of a binary classification 3D DenseNet121 to extract the relevant
dosimetric information directly from the 3D mandible radiation dose
distribution maps and predict the incidence of ORN. We compare the results to a
Random Forest ensemble with DVH-based parameters. Results. The 3D DenseNet121
model was able to discriminate ORN vs. non-ORN cases with an average AUC of
0.71 (0.64-0.79), compared to 0.65 (0.57-0.73) for the RF model. Conclusion.
Obtaining the dosimetric information directly from the clinical radiation dose
distribution maps may enhance the performance and functionality of ORN normal
tissue complication probability (NTCP) models.
Current artificial intelligence (AI) algorithms for short-axis cardiac magnetic resonance (CMR) segmentation achieve human performance for slices situated in the middle of the heart. However, an often-overlooked fact is that segmentation of the basal and apical slices is more difficult. During manual analysis, differences in the basal segmentations have been reported as one of the major sources of disagreement in human interobserver variability. In this work, we aim to investigate the performance of AI algorithms in segmenting basal and apical slices and design strategies to improve their segmentation. We trained all our models on a large dataset of clinical CMR studies obtained from two NHS hospitals (n=4,228) and evaluated them against two external datasets: ACDC (n=100) and M&Ms (n=321). Using manual segmentations as a reference, CMR slices were assigned to one of four regions: non-cardiac, base, middle, and apex. Using the nnU-Net framework as a baseline, we investigated two different approaches to reduce the segmentation performance gap between cardiac regions: (1) non-uniform batch sampling, which allows us to choose how often images from different regions are seen during training; and (2) a cardiac-region classification model followed by three (i.e. base, middle, and apex) region-specific segmentation models. We show that the classification and segmentation approach was best at reducing the performance gap across all datasets. We also show that improvements in the classification performance can subsequently lead to a significantly better performance in the segmentation task.
Fetal ultrasound screening during pregnancy plays a vital role in the early
detection of fetal malformations which have potential long-term health impacts.
The level of skill required to diagnose such malformations from live ultrasound
during examination is high and resources for screening are often limited. We
present an interpretable, atlas-learning segmentation method for automatic
diagnosis of Hypo-plastic Left Heart Syndrome (HLHS) from a single `4 Chamber
Heart' view image. We propose to extend the recently introduced
Image-and-Spatial Transformer Networks (Atlas-ISTN) into a framework that
enables sensitising atlas generation to disease. In this framework we can
jointly learn image segmentation, registration, atlas construction and disease
prediction while providing a maximum level of clinical interpretability
compared to direct image classification methods. As a result our segmentation
allows diagnoses competitive with expert-derived manual diagnosis and yields an
AUC-ROC of 0.978 (1043 cases for training, 260 for validation and 325 for
testing).
A variety of optimal control, estimation, system identification and design
problems can be formulated as functional optimization problems with
differential equality and inequality constraints. Since these problems are
infinite-dimensional and often do not have a known analytical solution, one has
to resort to numerical methods to compute an approximate solution. This paper
uses a unifying notation to outline some of the techniques used in the
transcription step of simultaneous direct methods (which
discretize-then-optimize) for solving continuous-time dynamic optimization
problems. We focus on collocation, integrated residual and Runge-Kutta schemes.
These transcription methods are then applied to a simulation case study to
answer a question that arose during the COVID-19 pandemic, namely: If there are
not enough ventilators, is it possible to ventilate more than one patient on a
single ventilator? The results suggest that it is possible, in principle, to
estimate individual patient parameters sufficiently accurately, using a
relatively small number of flow rate measurements, without needing to
disconnect a patient from the system or needing more than one flow rate sensor.
We also show that it is possible to ensure that two different patients can
indeed receive their desired tidal volume, by modifying the resistance
experienced by the air flow to each patient and controlling the ventilator
pressure.
Electronic Health Records are large repositories of valuable clinical data,
with a significant portion stored in unstructured text format. This textual
data includes clinical events (e.g., disorders, symptoms, findings, medications
and procedures) in context that if extracted accurately at scale can unlock
valuable downstream applications such as disease prediction. Using an existing
Named Entity Recognition and Linking methodology, MedCAT, these identified
concepts need to be further classified (contextualised) for their relevance to
the patient, and their temporal and negated status for example, to be useful
downstream. This study performs a comparative analysis of various natural
language models for medical text classification. Extensive experimentation
reveals the effectiveness of transformer-based language models, particularly
BERT. When combined with class imbalance mitigation techniques, BERT
outperforms Bi-LSTM models by up to 28% and the baseline BERT model by up to
16% for recall of the minority classes. The method has been implemented as part
of CogStack/MedCAT framework and made available to the community for further
research.
Splenomegaly, the enlargement of the spleen, is an important clinical indicator for various associated medical conditions, such as sickle cell disease (SCD). Spleen length measured from 2D ultrasound is the most widely used metric for characterising spleen size. However, it is still considered a surrogate measure, and spleen volume remains the gold standard for assessing spleen size. Accurate spleen volume measurement typically requires 3D imaging modalities, such as computed tomography or magnetic resonance imaging, but these are not widely available, especially in the Global South which has a high prevalence of SCD. In this work, we introduce a deep learning pipeline, DeepSPV, for precise spleen volume estimation from single or dual 2D ultrasound images. The pipeline involves a segmentation network and a variational autoencoder for learning low-dimensional representations from the estimated segmentations. We investigate three approaches for spleen volume estimation and our best model achieves 86.62%/92.5% mean relative volume accuracy (MRVA) under single-view/dual-view settings, surpassing the performance of human experts. In addition, the pipeline can provide confidence intervals for the volume estimates as well as offering benefits in terms of interpretability, which further support clinicians in decision-making when identifying splenomegaly. We evaluate the full pipeline using a highly realistic synthetic dataset generated by a diffusion model, achieving an overall MRVA of 83.0% from a single 2D ultrasound image. Our proposed DeepSPV is the first work to use deep learning to estimate 3D spleen volume from 2D ultrasound images and can be seamlessly integrated into the current clinical workflow for spleen assessment.
There are no more papers matching your filters at the moment.