Peking University Third Hospital
We introduce ColaCare, a framework that enhances Electronic Health Record
(EHR) modeling through multi-agent collaboration driven by Large Language
Models (LLMs). Our approach seamlessly integrates domain-specific expert models
with LLMs to bridge the gap between structured EHR data and text-based
reasoning. Inspired by the Multidisciplinary Team (MDT) approach used in
clinical settings, ColaCare employs two types of agents: DoctorAgents and a
MetaAgent, which collaboratively analyze patient data. Expert models process
and generate predictions from numerical EHR data, while LLM agents produce
reasoning references and decision-making reports within the MDT-driven
collaborative consultation framework. The MetaAgent orchestrates the
discussion, facilitating consultations and evidence-based debates among
DoctorAgents, simulating diverse expertise in clinical decision-making. We
additionally incorporate the Merck Manual of Diagnosis and Therapy (MSD)
medical guideline within a retrieval-augmented generation (RAG) module for
medical evidence support, addressing the challenge of knowledge currency.
Extensive experiments conducted on three EHR datasets demonstrate ColaCare's
superior performance in clinical mortality outcome and readmission prediction
tasks, underscoring its potential to revolutionize clinical decision support
systems and advance personalized precision medicine. All code, case studies and
a questionnaire are available at the project website:
this https URL
 University of Illinois at Urbana-Champaign
University of Illinois at Urbana-Champaign UC Berkeley
UC Berkeley NVIDIA
NVIDIA Johns Hopkins UniversityUniversity of BolognaJohns Hopkins School of MedicineIstituto Italiano di TecnologiaJagiellonian UniversityUniversity of California San FranciscoPeking University Third HospitalUniversity of Warmia and MazuryDiagnostic and Treatment Center Gammed
Johns Hopkins UniversityUniversity of BolognaJohns Hopkins School of MedicineIstituto Italiano di TecnologiaJagiellonian UniversityUniversity of California San FranciscoPeking University Third HospitalUniversity of Warmia and MazuryDiagnostic and Treatment Center GammedPanTS is a large-scale, multi-institutional dataset curated to advance research in pancreatic CT analysis. It contains 36,390 CT scans from 145 medical centers, with expert-validated, voxel-wise annotations of over 993,000 anatomical structures, covering pancreatic tumors, pancreas head, body, and tail, and 24 surrounding anatomical structures such as vascular/skeletal structures and abdominal/thoracic organs. Each scan includes metadata such as patient age, sex, diagnosis, contrast phase, in-plane spacing, slice thickness, etc. AI models trained on PanTS achieve significantly better performance in pancreatic tumor detection, localization, and segmentation compared to those trained on existing public datasets. Our analysis indicates that these gains are directly attributable to the 16x larger-scale tumor annotations and indirectly supported by the 24 additional surrounding anatomical structures. As the largest and most comprehensive resource of its kind, PanTS offers a new benchmark for developing and evaluating AI models in pancreatic CT analysis.
11 Oct 2025
MedAgentAudit: Diagnosing and Quantifying Collaborative Failure Modes in Medical Multi-Agent Systems
MedAgentAudit: Diagnosing and Quantifying Collaborative Failure Modes in Medical Multi-Agent Systems
While large language model (LLM)-based multi-agent systems show promise in simulating medical consultations, their evaluation is often confined to final-answer accuracy. This practice treats their internal collaborative processes as opaque "black boxes" and overlooks a critical question: is a diagnostic conclusion reached through a sound and verifiable reasoning pathway? The inscrutable nature of these systems poses a significant risk in high-stakes medical applications, potentially leading to flawed or untrustworthy conclusions. To address this, we conduct a large-scale empirical study of 3,600 cases from six medical datasets and six representative multi-agent frameworks. Through a rigorous, mixed-methods approach combining qualitative analysis with quantitative auditing, we develop a comprehensive taxonomy of collaborative failure modes. Our quantitative audit reveals four dominant failure patterns: flawed consensus driven by shared model deficiencies, suppression of correct minority opinions, ineffective discussion dynamics, and critical information loss during synthesis. This study demonstrates that high accuracy alone is an insufficient measure of clinical or public trust. It highlights the urgent need for transparent and auditable reasoning processes, a cornerstone for the responsible development and deployment of medical AI.
Large Language Models (LLMs) are revolutionizing bioinformatics, enabling
advanced analysis of DNA, RNA, proteins, and single-cell data. This survey
provides a systematic review of recent advancements, focusing on genomic
sequence modeling, RNA structure prediction, protein function inference, and
single-cell transcriptomics. Meanwhile, we also discuss several key challenges,
including data scarcity, computational complexity, and cross-omics integration,
and explore future directions such as multimodal learning, hybrid AI models,
and clinical applications. By offering a comprehensive perspective, this paper
underscores the transformative potential of LLMs in driving innovations in
bioinformatics and precision medicine.
In the field of medical decision-making, precise anomaly detection in medical imaging plays a pivotal role in aiding clinicians. However, previous work is reliant on large-scale datasets for training anomaly detection models, which increases the development cost. This paper first focuses on the task of medical image anomaly detection in the few-shot setting, which is critically significant for the medical field where data collection and annotation are both very expensive. We propose an innovative approach, MediCLIP, which adapts the CLIP model to few-shot medical image anomaly detection through self-supervised fine-tuning. Although CLIP, as a vision-language model, demonstrates outstanding zero-/fewshot performance on various downstream tasks, it still falls short in the anomaly detection of medical images. To address this, we design a series of medical image anomaly synthesis tasks to simulate common disease patterns in medical imaging, transferring the powerful generalization capabilities of CLIP to the task of medical image anomaly detection. When only few-shot normal medical images are provided, MediCLIP achieves state-of-the-art performance in anomaly detection and location compared to other methods. Extensive experiments on three distinct medical anomaly detection tasks have demonstrated the superiority of our approach. The code is available at this https URL.
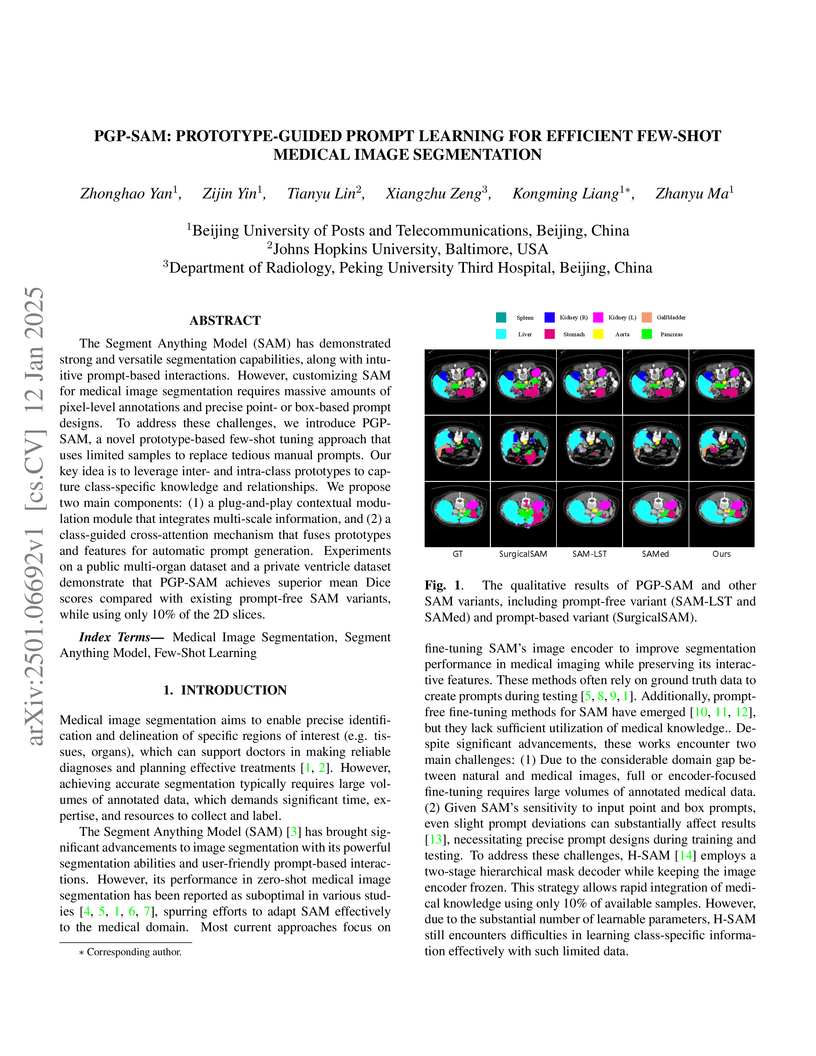
The Segment Anything Model (SAM) has demonstrated strong and versatile segmentation capabilities, along with intuitive prompt-based interactions. However, customizing SAM for medical image segmentation requires massive amounts of pixel-level annotations and precise point- or box-based prompt designs. To address these challenges, we introduce PGP-SAM, a novel prototype-based few-shot tuning approach that uses limited samples to replace tedious manual prompts. Our key idea is to leverage inter- and intra-class prototypes to capture class-specific knowledge and relationships. We propose two main components: (1) a plug-and-play contextual modulation module that integrates multi-scale information, and (2) a class-guided cross-attention mechanism that fuses prototypes and features for automatic prompt generation. Experiments on a public multi-organ dataset and a private ventricle dataset demonstrate that PGP-SAM achieves superior mean Dice scores compared with existing prompt-free SAM variants, while using only 10\% of the 2D slices.
Tissue mechanics--stiffness, density and impedance contrast--are broadly informative biomarkers across diseases, yet routine CT, MRI, and B-mode ultrasound rarely quantify them directly. While ultrasound tomography (UT) is intrinsically suited to in-vivo biomechanical assessment by capturing transmitted and reflected wavefields, efficient and accurate full-wave scattering models remain a bottleneck. Here, we introduce a generative neural physics framework that fuses generative models with physics-informed partial differential equation (PDE) solvers to produce rapid, high-fidelity 3D quantitative imaging of tissue mechanics. A compact neural surrogate for full-wave propagation is trained on limited cross-modality data, preserving physical accuracy while enabling efficient inversion. This enables, for the first time, accurate and efficient quantitative volumetric imaging of in vivo human breast and musculoskeletal tissues in under ten minutes, providing spatial maps of tissue mechanical properties not available from conventional reflection-mode or standard UT reconstructions. The resulting images reveal biomechanical features in bone, muscle, fat, and glandular tissues, maintaining structural resolution comparable to 3T MRI while providing substantially greater sensitivity to disease-related tissue mechanics.
Objective: The clinical diagnosis of developmental dysplasia of the hip (DDH) typically involves manually measuring key radiological angles -- Center-Edge (CE), Tonnis, and Sharp angles -- from pelvic radiographs, a process that is time-consuming and susceptible to variability. This study aims to develop an automated system that integrates these measurements to enhance the accuracy and consistency of DDH diagnosis.
Methods and procedures: We developed an end-to-end deep learning model for keypoint detection that accurately identifies eight anatomical keypoints from pelvic radiographs, enabling the automated calculation of CE, Tonnis, and Sharp angles. To support the diagnostic decision, we introduced a novel data-driven scoring system that combines the information from all three angles into a comprehensive and explainable diagnostic output.
Results: The system demonstrated superior consistency in angle measurements compared to a cohort of eight moderately experienced orthopedists. The intraclass correlation coefficients for the CE, Tonnis, and Sharp angles were 0.957 (95% CI: 0.952--0.962), 0.942 (95% CI: 0.937--0.947), and 0.966 (95% CI: 0.964--0.968), respectively. The system achieved a diagnostic F1 score of 0.863 (95% CI: 0.851--0.876), significantly outperforming the orthopedist group (0.777, 95% CI: 0.737--0.817, p = 0.005), as well as using clinical diagnostic criteria for each angle individually (p<0.001).
Conclusion: The proposed system provides reliable and consistent automated measurements of radiological angles and an explainable diagnostic output for DDH, outperforming moderately experienced clinicians.
Clinical impact: This AI-powered solution reduces the variability and potential errors of manual measurements, offering clinicians a more consistent and interpretable tool for DDH diagnosis.
Traditional one-shot medical image segmentation (MIS) methods use
registration networks to propagate labels from a reference atlas or rely on
comprehensive sampling strategies to generate synthetic labeled data for
training. However, these methods often struggle with registration errors and
low-quality synthetic images, leading to poor performance and generalization.
To overcome this, we introduce a novel one-shot MIS framework based on
knowledge distillation, which allows the network to directly 'see' real images
through a distillation process guided by image reconstruction. It focuses on
anatomical structures in a single labeled image and a few unlabeled ones. A
registration-based data augmentation network creates realistic, labeled
samples, while a feature distillation module helps the student network learn
segmentation from these samples, guided by the teacher network. During
inference, the streamlined student network accurately segments new images.
Evaluations on three public datasets (OASIS for T1 brain MRI, BCV for abdomen
CT, and VerSe for vertebrae CT) show superior segmentation performance and
generalization across different medical image datasets and modalities compared
to leading methods. Our code is available at
this https URL
Objective: Peritoneal Dialysis (PD) is one of the most widely used life-supporting therapies for patients with End-Stage Renal Disease (ESRD). Predicting mortality risk and identifying modifiable risk factors based on the Electronic Medical Records (EMR) collected along with the follow-up visits are of great importance for personalized medicine and early intervention. Here, our objective is to develop a deep learning model for a real-time, individualized, and interpretable mortality prediction model - AICare. Method and Materials: Our proposed model consists of a multi-channel feature extraction module and an adaptive feature importance recalibration module. AICare explicitly identifies the key features that strongly indicate the outcome prediction for each patient to build the health status embedding individually. This study has collected 13,091 clinical follow-up visits and demographic data of 656 PD patients. To verify the application universality, this study has also collected 4,789 visits of 1,363 hemodialysis dialysis (HD) as an additional experiment dataset to test the prediction performance, which will be discussed in the Appendix. Results: 1) Experiment results show that AICare achieves 81.6%/74.3% AUROC and 47.2%/32.5% AUPRC for the 1-year mortality prediction task on PD/HD dataset respectively, which outperforms the state-of-the-art comparative deep learning models. 2) This study first provides a comprehensive elucidation of the relationship between the causes of mortality in patients with PD and clinical features based on an end-to-end deep learning model. 3) This study first reveals the pattern of variation in the importance of each feature in the mortality prediction based on built-in interpretability. 4) We develop a practical AI-Doctor interaction system to visualize the trajectory of patients' health status and risk indicators.
18 Mar 2024
In the field of medical image analysis, the scarcity of Chinese chest X-ray
report datasets has hindered the development of technology for generating
Chinese chest X-ray reports. On one hand, the construction of a Chinese chest
X-ray report dataset is limited by the time-consuming and costly process of
accurate expert disease annotation. On the other hand, a single natural
language generation metric is commonly used to evaluate the similarity between
generated and ground-truth reports, while the clinical accuracy and
effectiveness of the generated reports rely on an accurate disease labeler
(classifier). To address the issues, this study proposes a disease labeler
tailored for the generation of Chinese chest X-ray reports. This labeler
leverages a dual BERT architecture to handle diagnostic reports and clinical
information separately and constructs a hierarchical label learning algorithm
based on the affiliation between diseases and body parts to enhance text
classification performance. Utilizing this disease labeler, a Chinese chest
X-ray report dataset comprising 51,262 report samples was established. Finally,
experiments and analyses were conducted on a subset of expert-annotated Chinese
chest X-ray reports, validating the effectiveness of the proposed disease
labeler.
ADAptation is an unsupervised active learning framework that uses diffusion models to transform target breast ultrasound images into a source-like style, then applies hypersphere-constrained contrastive learning to select informative samples. This approach significantly reduces annotation costs, achieving a 0.8081 classification accuracy with only 20% labeled target data while consistently outperforming other active learning methods across diverse deep learning models and multi-center datasets.
The brain extracellular space (ECS), an irregular, extremely tortuous
nanoscale space located between cells or between cells and blood vessels, is
crucial for nerve cell survival. It plays a pivotal role in high-level brain
functions such as memory, emotion, and sensation. However, the specific form of
molecular transport within the ECS remain elusive. To address this challenge,
this paper proposes a novel approach to quantitatively analyze the molecular
transport within the ECS by solving an inverse problem derived from the
advection-diffusion equation (ADE) using a physics-informed neural network
(PINN). PINN provides a streamlined solution to the ADE without the need for
intricate mathematical formulations or grid settings. Additionally, the
optimization of PINN facilitates the automatic computation of the diffusion
coefficient governing long-term molecule transport and the velocity of
molecules driven by advection. Consequently, the proposed method allows for the
quantitative analysis and identification of the specific pattern of molecular
transport within the ECS through the calculation of the Peclet number.
Experimental validation on two datasets of magnetic resonance images (MRIs)
captured at different time points showcases the effectiveness of the proposed
method. Notably, our simulations reveal identical molecular transport patterns
between datasets representing rats with tracer injected into the same brain
region. These findings highlight the potential of PINN as a promising tool for
comprehensively exploring molecular transport within the ECS.
CarveMix introduces a simple, lesion-aware data augmentation method for 3D brain lesion segmentation, generating synthetic samples by combining lesion-centric regions from existing images with consistent annotations. This approach consistently improves segmentation accuracy, especially in low-data regimes, outperforming traditional and generic "mix"-based augmentation techniques on stroke lesion datasets.
Recent advances in automated radiology report generation from chest X-rays using deep learning algorithms have the potential to significantly reduce the arduous workload of radiologists. However, due to the inherent massive data bias in radiology images, where abnormalities are typically subtle and sparsely distributed, existing methods often produce fluent yet medically inaccurate reports, limiting their applicability in clinical practice. To address this issue effectively, we propose a Semantically Informed Salient Regions-guided (SISRNet) report generation method. Specifically, our approach explicitly identifies salient regions with medically critical characteristics using fine-grained cross-modal semantics. Then, SISRNet systematically focuses on these high-information regions during both image modeling and report generation, effectively capturing subtle abnormal findings, mitigating the negative impact of data bias, and ultimately generating clinically accurate reports. Compared to its peers, SISRNet demonstrates superior performance on widely used IU-Xray and MIMIC-CXR datasets.
Accurate multi-needle localization in intraoperative CT images is crucial for optimizing seed placement in pelvic seed implant brachytherapy. However, this task is challenging due to poor image contrast and needle adhesion. This paper presents a novel approach that reframes needle localization as a tip-handle detection and matching problem to overcome these difficulties. An anchor-free network, based on HRNet, is proposed to extract multi-scale features and accurately detect needle tips and handles by predicting their centers and orientations using decoupled branches for heatmap regression and polar angle prediction. To associate detected tips and handles into individual needles, a greedy matching and merging (GMM) method designed to solve the unbalanced assignment problem with constraints (UAP-C) is presented. The GMM method iteratively selects the most probable tip-handle pairs and merges them based on a distance metric to reconstruct 3D needle paths. Evaluated on a dataset of 100 patients, the proposed method demonstrates superior performance, achieving higher precision and F1 score compared to a segmentation-based method utilizing the nnUNet model,thereby offering a more robust and accurate solution for needle localization in complex clinical scenarios.
Automated brain lesion segmentation provides valuable information for the
analysis and intervention of patients. In particular, methods based on
convolutional neural networks (CNNs) have achieved state-of-the-art
segmentation performance. However, CNNs usually require a decent amount of
annotated data, which may be costly and time-consuming to obtain. Since
unannotated data is generally abundant, it is desirable to use unannotated data
to improve the segmentation performance for CNNs when limited annotated data is
available. In this work, we propose a semi-supervised learning (SSL) approach
to brain lesion segmentation, where unannotated data is incorporated into the
training of CNNs. We adapt the mean teacher model, which is originally
developed for SSL-based image classification, for brain lesion segmentation.
Assuming that the network should produce consistent outputs for similar inputs,
a loss of segmentation consistency is designed and integrated into a
self-ensembling framework. Specifically, we build a student model and a teacher
model, which share the same CNN architecture for segmentation. The student and
teacher models are updated alternately. At each step, the student model learns
from the teacher model by minimizing the weighted sum of the segmentation loss
computed from annotated data and the segmentation consistency loss between the
teacher and student models computed from unannotated data. Then, the teacher
model is updated by combining the updated student model with the historical
information of teacher models using an exponential moving average strategy. For
demonstration, the proposed approach was evaluated on ischemic stroke lesion
segmentation, where it improves stroke lesion segmentation with the
incorporation of unannotated data.
05 Dec 2021
Purpose: This study aims to develop a novel approach to extracting and measuring uncertain biomedical knowledge from scientific statements. Design/methodology/approach: Taking cardiovascular research publications in China as a sample, we extracted the SPO triples as knowledge unit and the hedging/conflicting uncertainties as the knowledge context. We introduced Information Entropy and Uncertainty Rate as potential metrics to quantity the uncertainty of biomedical knowledge claims represented at different levels, such as the SPO triples (micro level), as well as the semantic type pairs (micro-level). Findings: The results indicated that while the number of scientific publications and total SPO triples showed a liner growth, the novel SPO triples occurring per year remained stable. After examining the frequency of uncertain cue words in different part of scientific statements, we found hedging words tend to appear in conclusive and purposeful sentences, whereas conflicting terms often appear in background and act as the premise (e.g., unsettled scientific issues) of the work to be investigated. Practical implications: Our approach identified major uncertain knowledge areas, such as diagnostic biomarkers, genetic characteristics, and pharmacologic therapies surrounding cardiovascular diseases in China. These areas are suggested to be prioritized in which new hypotheses need to be verified, and disputes, conflicts, as well as contradictions to be settled further.
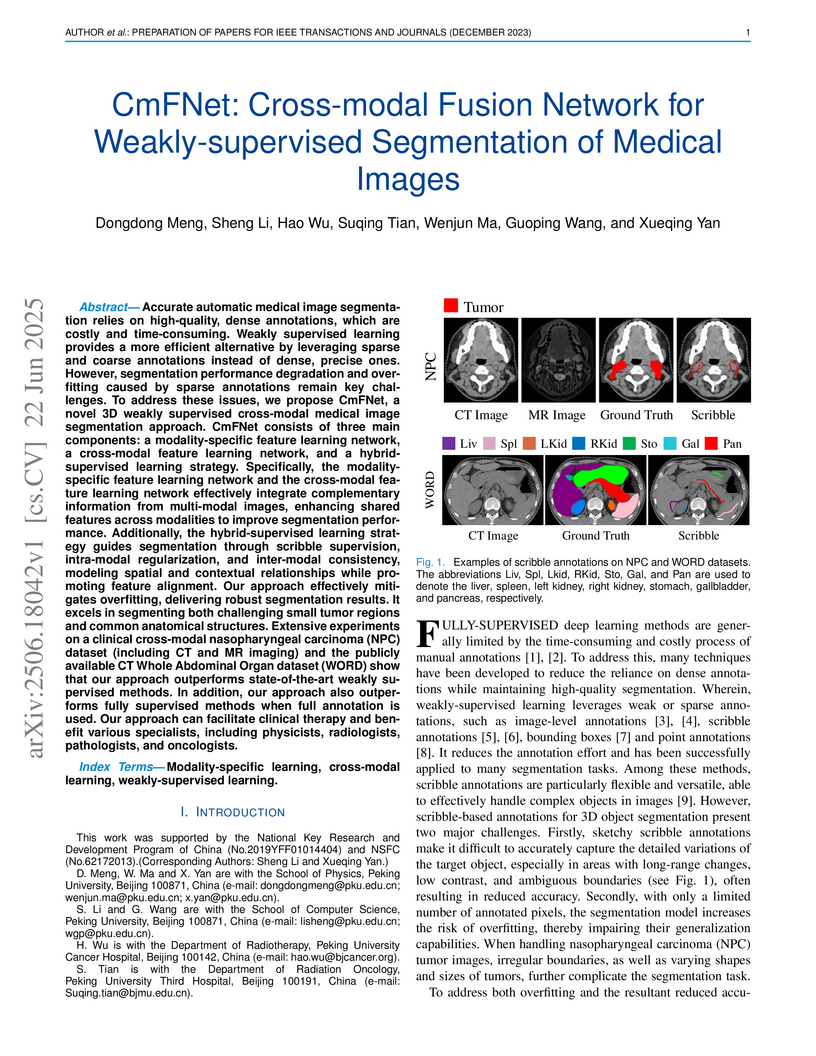
Accurate automatic medical image segmentation relies on high-quality, dense annotations, which are costly and time-consuming. Weakly supervised learning provides a more efficient alternative by leveraging sparse and coarse annotations instead of dense, precise ones. However, segmentation performance degradation and overfitting caused by sparse annotations remain key challenges. To address these issues, we propose CmFNet, a novel 3D weakly supervised cross-modal medical image segmentation approach. CmFNet consists of three main components: a modality-specific feature learning network, a cross-modal feature learning network, and a hybrid-supervised learning strategy. Specifically, the modality-specific feature learning network and the cross-modal feature learning network effectively integrate complementary information from multi-modal images, enhancing shared features across modalities to improve segmentation performance. Additionally, the hybrid-supervised learning strategy guides segmentation through scribble supervision, intra-modal regularization, and inter-modal consistency, modeling spatial and contextual relationships while promoting feature alignment. Our approach effectively mitigates overfitting, delivering robust segmentation results. It excels in segmenting both challenging small tumor regions and common anatomical structures. Extensive experiments on a clinical cross-modal nasopharyngeal carcinoma (NPC) dataset (including CT and MR imaging) and the publicly available CT Whole Abdominal Organ dataset (WORD) show that our approach outperforms state-of-the-art weakly supervised methods. In addition, our approach also outperforms fully supervised methods when full annotation is used. Our approach can facilitate clinical therapy and benefit various specialists, including physicists, radiologists, pathologists, and oncologists.
Segmentation of pathological images is essential for accurate disease
diagnosis. The quality of manual labels plays a critical role in segmentation
accuracy; yet, in practice, the labels between pathologists could be
inconsistent, thus confusing the training process. In this work, we propose a
novel label re-weighting framework to account for the reliability of different
experts' labels on each pixel according to its surrounding features. We further
devise a new attention heatmap, which takes roughness as prior knowledge to
guide the model to focus on important regions. Our approach is evaluated on the
public Gleason 2019 datasets. The results show that our approach effectively
improves the model's robustness against noisy labels and outperforms
state-of-the-art approaches.
There are no more papers matching your filters at the moment.