RigshospitaletCopenhagen University Hospital
The ethical and legal imperative to share research data without causing harm requires careful attention to privacy risks. While mounting evidence demonstrates that data sharing benefits science, legitimate concerns persist regarding the potential leakage of personal information that could lead to reidentification and subsequent harm. We reviewed metadata accompanying neuroimaging datasets from six heterogeneous studies openly available on OpenNeuro, involving participants across the lifespan, from children to older adults, with and without clinical diagnoses, and including associated clinical score data. Using metaprivBIDS (this https URL), a novel tool for the systematic assessment of privacy in tabular data, we found that privacy is generally well maintained, with serious vulnerabilities being rare. Nonetheless, minor issues were identified in nearly all datasets and warrant mitigation. Notably, clinical score data (e.g., neuropsychological results) posed minimal reidentification risk, whereas demographic variables (age, sex, race, income, and geolocation) represented the principal privacy vulnerabilities. We outline practical measures to address these risks, enabling safer data sharing practices.
Humans are highly dependent on the ability to process audio in order to
interact through conversation and navigate from sound. For this, the shape of
the ear acts as a mechanical audio filter. The anatomy of the outer human ear
canal to approximately 15-20 mm beyond the Tragus is well described because of
its importance for customized hearing aid production. This is however not the
case for the part of the ear canal that is embedded in the skull, until the
typanic membrane. Due to the sensitivity of the outer ear, this part, referred
to as the bony part, has only been described in a few population studies and
only ex-vivo. We present a study of the entire ear canal including the bony
part and the tympanic membrane. We form an average ear canal from a number of
MRI scans using standard image registration methods. We show that the obtained
representation is realistic in the sense that it has acoustical properties
almost identical to a real ear.
Large language models (LLMs) such as GPT-4 have recently demonstrated impressive results across a wide range of tasks. LLMs are still limited, however, in that they frequently fail at complex reasoning, their reasoning processes are opaque, they are prone to 'hallucinate' facts, and there are concerns about their underlying biases. Letting models verbalize reasoning steps as natural language, a technique known as chain-of-thought prompting, has recently been proposed as a way to address some of these issues. Here we present ThoughtSource, a meta-dataset and software library for chain-of-thought (CoT) reasoning. The goal of ThoughtSource is to improve future artificial intelligence systems by facilitating qualitative understanding of CoTs, enabling empirical evaluations, and providing training data. This first release of ThoughtSource integrates seven scientific/medical, three general-domain and five math word question answering datasets.
Curvilinear structure segmentation is important in medical imaging,
quantifying structures such as vessels, airways, neurons, or organ boundaries
in 2D slices. Segmentation via pixel-wise classification often fails to capture
the small and low-contrast curvilinear structures. Prior topological
information is typically used to address this problem, often at an expensive
computational cost, and sometimes requiring prior knowledge of the expected
topology.
We present DTU-Net, a data-driven approach to topology-preserving curvilinear
structure segmentation. DTU-Net consists of two sequential, lightweight U-Nets,
dedicated to texture and topology, respectively. While the texture net makes a
coarse prediction using image texture information, the topology net learns
topological information from the coarse prediction by employing a triplet loss
trained to recognize false and missed splits in the structure. We conduct
experiments on a challenging multi-class ultrasound scan segmentation dataset
as well as a well-known retinal imaging dataset. Results show that our model
outperforms existing approaches in both pixel-wise segmentation accuracy and
topological continuity, with no need for prior topological knowledge.
Radiology reports contain rich clinical information that can be used to train imaging models without relying on costly manual annotation. However, existing approaches face critical limitations: rule-based methods struggle with linguistic variability, supervised models require large annotated datasets, and recent LLM-based systems depend on closed-source or resource-intensive models that are unsuitable for clinical use. Moreover, current solutions are largely restricted to English and single-modality, single-taxonomy datasets. We introduce MOSAIC, a multilingual, taxonomy-agnostic, and computationally efficient approach for radiological report classification. Built on a compact open-access language model (MedGemma-4B), MOSAIC supports both zero-/few-shot prompting and lightweight fine-tuning, enabling deployment on consumer-grade GPUs. We evaluate MOSAIC across seven datasets in English, Spanish, French, and Danish, spanning multiple imaging modalities and label taxonomies. The model achieves a mean macro F1 score of 88 across five chest X-ray datasets, approaching or exceeding expert-level performance, while requiring only 24 GB of GPU memory. With data augmentation, as few as 80 annotated samples are sufficient to reach a weighted F1 score of 82 on Danish reports, compared to 86 with the full 1600-sample training set. MOSAIC offers a practical alternative to large or proprietary LLMs in clinical settings. Code and models are open-source. We invite the community to evaluate and extend MOSAIC on new languages, taxonomies, and modalities.
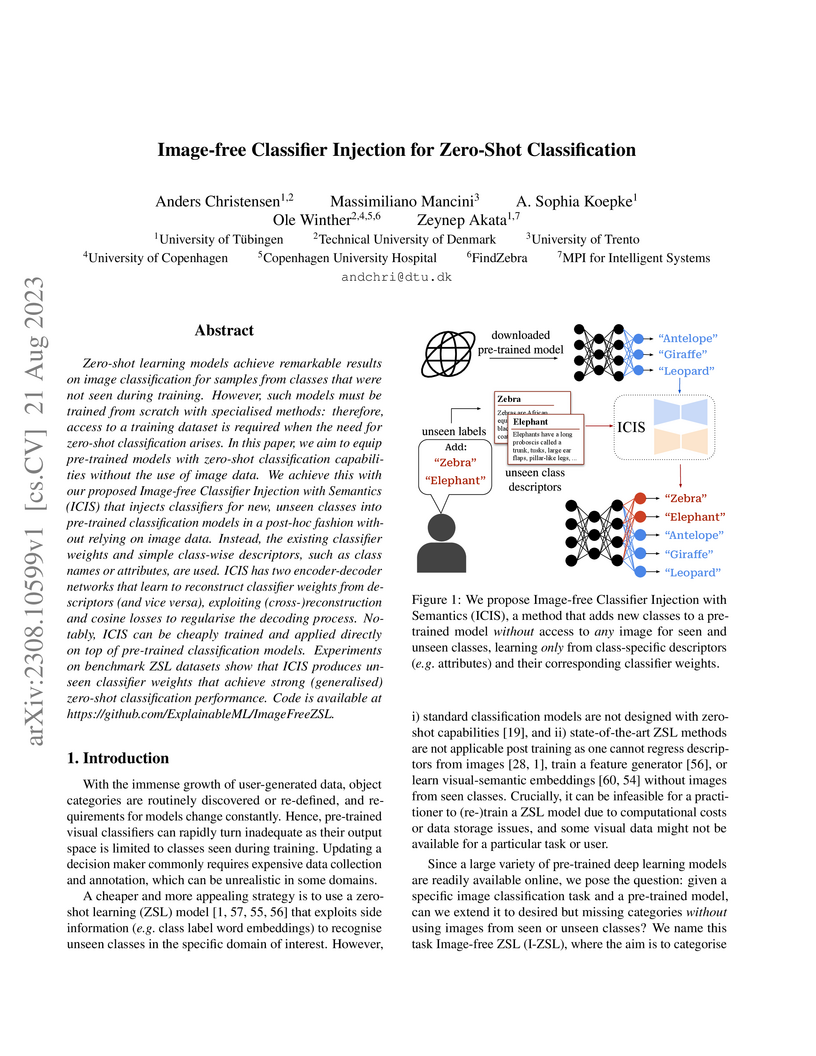
Zero-shot learning models achieve remarkable results on image classification for samples from classes that were not seen during training. However, such models must be trained from scratch with specialised methods: therefore, access to a training dataset is required when the need for zero-shot classification arises. In this paper, we aim to equip pre-trained models with zero-shot classification capabilities without the use of image data. We achieve this with our proposed Image-free Classifier Injection with Semantics (ICIS) that injects classifiers for new, unseen classes into pre-trained classification models in a post-hoc fashion without relying on image data. Instead, the existing classifier weights and simple class-wise descriptors, such as class names or attributes, are used. ICIS has two encoder-decoder networks that learn to reconstruct classifier weights from descriptors (and vice versa), exploiting (cross-)reconstruction and cosine losses to regularise the decoding process. Notably, ICIS can be cheaply trained and applied directly on top of pre-trained classification models. Experiments on benchmark ZSL datasets show that ICIS produces unseen classifier weights that achieve strong (generalised) zero-shot classification performance. Code is available at this https URL .
Heidelberg University Stanford UniversityIT University of Copenhagen
Stanford UniversityIT University of Copenhagen University of CopenhagenCONICET
University of CopenhagenCONICET Emory UniversityAarhus UniversityThe Hebrew University of JerusalemUniversitat de BarcelonaTechnical University of Denmark
Emory UniversityAarhus UniversityThe Hebrew University of JerusalemUniversitat de BarcelonaTechnical University of Denmark University of GroningenUniversity of Southern DenmarkRadboud University Medical CenterGerman Cancer Research CenterRigshospitaletARTORG Center for Biomedical Engineering Research, University of BernUniversity of Buenos AiresHelmholtz ImagingLunitFederal University of Espírito SantoCopenhagen University Hospital, Herlev and GentofteCerebriu A/SChurchill Hospital, Oxford University HospitalsSteno Diabetes Center Aarhus, Aarhus University HospitalUniversity Hospital Bern, University of BernEurécom
University of GroningenUniversity of Southern DenmarkRadboud University Medical CenterGerman Cancer Research CenterRigshospitaletARTORG Center for Biomedical Engineering Research, University of BernUniversity of Buenos AiresHelmholtz ImagingLunitFederal University of Espírito SantoCopenhagen University Hospital, Herlev and GentofteCerebriu A/SChurchill Hospital, Oxford University HospitalsSteno Diabetes Center Aarhus, Aarhus University HospitalUniversity Hospital Bern, University of BernEurécom
 Stanford UniversityIT University of Copenhagen
Stanford UniversityIT University of Copenhagen University of CopenhagenCONICET
University of CopenhagenCONICET Emory UniversityAarhus UniversityThe Hebrew University of JerusalemUniversitat de BarcelonaTechnical University of Denmark
Emory UniversityAarhus UniversityThe Hebrew University of JerusalemUniversitat de BarcelonaTechnical University of Denmark University of GroningenUniversity of Southern DenmarkRadboud University Medical CenterGerman Cancer Research CenterRigshospitaletARTORG Center for Biomedical Engineering Research, University of BernUniversity of Buenos AiresHelmholtz ImagingLunitFederal University of Espírito SantoCopenhagen University Hospital, Herlev and GentofteCerebriu A/SChurchill Hospital, Oxford University HospitalsSteno Diabetes Center Aarhus, Aarhus University HospitalUniversity Hospital Bern, University of BernEurécom
University of GroningenUniversity of Southern DenmarkRadboud University Medical CenterGerman Cancer Research CenterRigshospitaletARTORG Center for Biomedical Engineering Research, University of BernUniversity of Buenos AiresHelmholtz ImagingLunitFederal University of Espírito SantoCopenhagen University Hospital, Herlev and GentofteCerebriu A/SChurchill Hospital, Oxford University HospitalsSteno Diabetes Center Aarhus, Aarhus University HospitalUniversity Hospital Bern, University of BernEurécomDatasets play a critical role in medical imaging research, yet issues such as label quality, shortcuts, and metadata are often overlooked. This lack of attention may harm the generalizability of algorithms and, consequently, negatively impact patient outcomes. While existing medical imaging literature reviews mostly focus on machine learning (ML) methods, with only a few focusing on datasets for specific applications, these reviews remain static -- they are published once and not updated thereafter. This fails to account for emerging evidence, such as biases, shortcuts, and additional annotations that other researchers may contribute after the dataset is published. We refer to these newly discovered findings of datasets as research artifacts. To address this gap, we propose a living review that continuously tracks public datasets and their associated research artifacts across multiple medical imaging applications. Our approach includes a framework for the living review to monitor data documentation artifacts, and an SQL database to visualize the citation relationships between research artifact and dataset. Lastly, we discuss key considerations for creating medical imaging datasets, review best practices for data annotation, discuss the significance of shortcuts and demographic diversity, and emphasize the importance of managing datasets throughout their entire lifecycle. Our demo is publicly available at this http URL.
Diffusion models may be viewed as hierarchical variational autoencoders (VAEs) with two improvements: parameter sharing for the conditional distributions in the generative process and efficient computation of the loss as independent terms over the hierarchy. We consider two changes to the diffusion model that retain these advantages while adding flexibility to the model. Firstly, we introduce a data- and depth-dependent mean function in the diffusion process, which leads to a modified diffusion loss. Our proposed framework, DiffEnc, achieves a statistically significant improvement in likelihood on CIFAR-10. Secondly, we let the ratio of the noise variance of the reverse encoder process and the generative process be a free weight parameter rather than being fixed to 1. This leads to theoretical insights: For a finite depth hierarchy, the evidence lower bound (ELBO) can be used as an objective for a weighted diffusion loss approach and for optimizing the noise schedule specifically for inference. For the infinite-depth hierarchy, on the other hand, the weight parameter has to be 1 to have a well-defined ELBO.
Domain randomization through synthesis is a powerful strategy to train networks that are unbiased with respect to the domain of the input images. Randomization allows networks to see a virtually infinite range of intensities and artifacts during training, thereby minimizing overfitting to appearance and maximizing generalization to unseen data. Although powerful, this approach relies on the accurate tuning of a large set of hyperparameters that govern the probabilistic distribution of the synthesized images. Instead of manually tuning these parameters, we introduce Learn2Synth, a novel procedure in which synthesis parameters are learned using a small set of real labeled data. Unlike methods that impose constraints to align synthetic data with real data (e.g., contrastive or adversarial techniques), which risk misaligning the image and its label map, we tune an augmentation engine such that a segmentation network trained on synthetic data has optimal accuracy when applied to real data. This approach allows the training procedure to benefit from real labeled examples, without ever using these real examples to train the segmentation network, which avoids biasing the network towards the properties of the training set. Specifically, we develop parametric and nonparametric strategies to enhance synthetic images in a way that improves the performance of the segmentation network. We demonstrate the effectiveness of this learning strategy on synthetic and real-world brain scans. Code is available at: this https URL.
A large-scale heterogeneous 3D magnetic resonance brain imaging dataset for self-supervised learning
A large-scale heterogeneous 3D magnetic resonance brain imaging dataset for self-supervised learning
We present FOMO60K, a large-scale, heterogeneous dataset of 60,529 brain
Magnetic Resonance Imaging (MRI) scans from 13,900 sessions and 11,187
subjects, aggregated from 16 publicly available sources. The dataset includes
both clinical- and research-grade images, multiple MRI sequences, and a wide
range of anatomical and pathological variability, including scans with large
brain anomalies. Minimal preprocessing was applied to preserve the original
image characteristics while reducing barriers to entry for new users.
Accompanying code for self-supervised pretraining and finetuning is provided.
FOMO60K is intended to support the development and benchmarking of
self-supervised learning methods in medical imaging at scale.
Medical imaging models have been shown to encode information about patient
demographics such as age, race, and sex in their latent representation, raising
concerns about their potential for discrimination. Here, we ask whether
requiring models not to encode demographic attributes is desirable. We point
out that marginal and class-conditional representation invariance imply the
standard group fairness notions of demographic parity and equalized odds,
respectively. In addition, however, they require matching the risk
distributions, thus potentially equalizing away important group differences.
Enforcing the traditional fairness notions directly instead does not entail
these strong constraints. Moreover, representationally invariant models may
still take demographic attributes into account for deriving predictions,
implying unequal treatment - in fact, achieving representation invariance may
require doing so. In theory, this can be prevented using counterfactual notions
of (individual) fairness or invariance. We caution, however, that properly
defining medical image counterfactuals with respect to demographic attributes
is fraught with challenges. Finally, we posit that encoding demographic
attributes may even be advantageous if it enables learning a task-specific
encoding of demographic features that does not rely on social constructs such
as 'race' and 'gender.' We conclude that demographically invariant
representations are neither necessary nor sufficient for fairness in medical
imaging. Models may need to encode demographic attributes, lending further
urgency to calls for comprehensive model fairness assessments in terms of
predictive performance across diverse patient groups.
The rhythmic pumping motion of the heart stands as a cornerstone in life, as it circulates blood to the entire human body through a series of carefully timed contractions of the individual chambers. Changes in the size, shape and movement of the chambers can be important markers for cardiac disease and modeling this in relation to clinical demography or disease is therefore of interest. Existing methods for spatio-temporal modeling of the human heart require shape correspondence over time or suffer from large memory requirements, making it difficult to use for complex anatomies. We introduce a novel conditional generative model, where the shape and movement is modeled implicitly in the form of a spatio-temporal neural distance field and conditioned on clinical demography. The model is based on an auto-decoder architecture and aims to disentangle the individual variations from that related to the clinical demography. It is tested on the left atrium (including the left atrial appendage), where it outperforms current state-of-the-art methods for anatomical sequence completion and generates synthetic sequences that realistically mimics the shape and motion of the real left atrium. In practice, this means we can infer functional measurements from a static image, generate synthetic populations with specified demography or disease and investigate how non-imaging clinical data effect the shape and motion of cardiac anatomies.
Medical image segmentation is often considered as the task of labelling each
pixel or voxel as being inside or outside a given anatomy. Processing the
images at their original size and resolution often result in insuperable memory
requirements, but downsampling the images leads to a loss of important details.
Instead of aiming to represent a smooth and continuous surface in a binary
voxel-grid, we propose to learn a Neural Unsigned Distance Field (NUDF)
directly from the image. The small memory requirements of NUDF allow for high
resolution processing, while the continuous nature of the distance field allows
us to create high resolution 3D mesh models of shapes of any topology (i.e.
open surfaces). We evaluate our method on the task of left atrial appendage
(LAA) segmentation from Computed Tomography (CT) images. The LAA is a complex
and highly variable shape, being thus difficult to represent with traditional
segmentation methods using discrete labelmaps. With our proposed method, we are
able to predict 3D mesh models that capture the details of the LAA and achieve
accuracy in the order of the voxel spacing in the CT images.
Spherical or omni-directional images offer an immersive visual format appealing to a wide range of computer vision applications. However, geometric properties of spherical images pose a major challenge for models and metrics designed for ordinary 2D images. Here, we show that direct application of Fréchet Inception Distance (FID) is insufficient for quantifying geometric fidelity in spherical images. We introduce two quantitative metrics accounting for geometric constraints, namely Omnidirectional FID (OmniFID) and Discontinuity Score (DS). OmniFID is an extension of FID tailored to additionally capture field-of-view requirements of the spherical format by leveraging cubemap projections. DS is a kernel-based seam alignment score of continuity across borders of 2D representations of spherical images. In experiments, OmniFID and DS quantify geometry fidelity issues that are undetected by FID.
The research by Gopinath et al. at Massachusetts General Hospital introduces "recon-any," a deep learning framework designed to enable robust cortical surface reconstruction and morphometric analysis from low-field MRI (LF-MRI) scans. The method achieves high accuracy for surface area and gray matter volume measurements, extending advanced neuroimaging capabilities to portable and postmortem imaging applications.
25 Nov 2024
In recent years, the use of deep learning (DL) methods, including convolutional neural networks (CNNs) and vision transformers (ViTs), has significantly advanced computational pathology, enhancing both diagnostic accuracy and efficiency. Hematoxylin and Eosin (H&E) Whole Slide Images (WSI) plays a crucial role by providing detailed tissue samples for the analysis and training of DL models. However, WSIs often contain regions with artifacts such as tissue folds, blurring, as well as non-tissue regions (background), which can negatively impact DL model performance. These artifacts are diagnostically irrelevant and can lead to inaccurate results. This paper proposes a fully automatic supervised DL pipeline for WSI Quality Assessment (WSI-QA) that uses a fused model combining CNNs and ViTs to detect and exclude WSI regions with artifacts, ensuring that only qualified WSI regions are used to build DL-based computational pathology applications. The proposed pipeline employs a pixel-based segmentation model to classify WSI regions as either qualified or non- qualified based on the presence of artifacts. The proposed model was trained on a large and diverse dataset and validated with internal and external data from various human organs, scanners, and H&E staining procedures. Quantitative and qualitative evaluations demonstrate the superiority of the proposed model, which outperforms state- of-the-art methods in WSI artifact detection. The proposed model consistently achieved over 95% accuracy, precision, recall, and F1 score across all artifact types. Furthermore, the WSI-QA pipeline shows strong generalization across different tissue types and scanning conditions.
Generative models such as Generative Adversarial Networks (GANs) and
Variational Autoencoders (VAEs) play an increasingly important role in medical
image analysis. The latent spaces of these models often show semantically
meaningful directions corresponding to human-interpretable image
transformations. However, until now, their exploration for medical images has
been limited due to the requirement of supervised data. Several methods for
unsupervised discovery of interpretable directions in GAN latent spaces have
shown interesting results on natural images. This work explores the potential
of applying these techniques on medical images by training a GAN and a VAE on
thoracic CT scans and using an unsupervised method to discover interpretable
directions in the resulting latent space. We find several directions
corresponding to non-trivial image transformations, such as rotation or breast
size. Furthermore, the directions show that the generative models capture 3D
structure despite being presented only with 2D data. The results show that
unsupervised methods to discover interpretable directions in GANs generalize to
VAEs and can be applied to medical images. This opens a wide array of future
work using these methods in medical image analysis.
Congenital malformations of the brain are among the most common fetal abnormalities that impact fetal development. Previous anomaly detection methods on ultrasound images are based on supervised learning, rely on manual annotations, and risk missing underrepresented categories. In this work, we frame fetal brain anomaly detection as an unsupervised task using diffusion models. To this end, we employ an inpainting-based Noise Agnostic Anomaly Detection approach that identifies the abnormality using diffusion-reconstructed fetal brain images from multiple noise levels. Our approach only requires normal fetal brain ultrasound images for training, addressing the limited availability of abnormal data. Our experiments on a real-world clinical dataset show the potential of using unsupervised methods for fetal brain anomaly detection. Additionally, we comprehensively evaluate how different noise types affect diffusion models in the fetal anomaly detection domain.
High-resolution colon segmentation is crucial for clinical and research
applications, such as digital twins and personalized medicine. However, the
leading open-source abdominal segmentation tool, TotalSegmentator, struggles
with accuracy for the colon, which has a complex and variable shape, requiring
time-intensive labeling. Here, we present the first fully automatic
high-resolution colon segmentation method. To develop it, we first created a
high resolution colon dataset using a pipeline that combines region growing
with interactive machine learning to efficiently and accurately label the colon
on CT colonography (CTC) images. Based on the generated dataset consisting of
435 labeled CTC images we trained an nnU-Net model for fully automatic colon
segmentation. Our fully automatic model achieved an average symmetric surface
distance of 0.2 mm (vs. 4.0 mm from TotalSegmentator) and a 95th percentile
Hausdorff distance of 1.0 mm (vs. 18 mm from TotalSegmentator). Our
segmentation accuracy substantially surpasses TotalSegmentator. We share our
trained model and pipeline code, providing the first and only open-source tool
for high-resolution colon segmentation. Additionally, we created a large-scale
dataset of publicly available high-resolution colon labels.
13 Dec 2023
OBJECTIVE: Assessing the biomechanical behaviour of the aortic wall in vivo
can potentially improve the diagnosis and prognosis of patients with abdominal
aortic aneurysms (AAA). With ultrasound, AAA wall stiffness can be estimated as
the diameter change in response to blood pressure. However, this measurement
has shown limited reproducibility, possibly influenced by the unknown effects
of the highly variable ultrasound probe pressure. This study addressed this gap
by analyzing time-resolved ultrasound sequences from AAA patients.METHOD:
Two-dimensional ultrasound sequences were acquired from AAA patients,
alternatively applying light and firm probe pressure. Diameter variations and
stiffness were evaluated and compared. An in silico simulation was performed to
support the in vivo observations.RESULTS: It was found that half of the AAAs
were particularly responsive to the probe pressure: in these patients, the
cyclic diameter variation between diastole and systole changed from 1% at light
probe pressure to 5% at firm probe pressure and the estimated stiffness
decreases by a factor of 6.3. Conversely, the other half of the patients were
less responsive: diameter variations increased marginally and stiffness
decreased by a factor of 1.5 when transitioning from light to firm probe
pressure. Through numerical simulations, we were able to show that the rather
counter-intuitive stiffness decrease observed in the responsive cohort is
actually consistent with a biomechanical model.CONCLUSION: We conclude that
probe pressure has an important effect on the estimation of AAA wall stiffness.
Ultimately, our findings suggest potential implications for AAA mechanical
characterization via ultrasound imaging.
There are no more papers matching your filters at the moment.