University of Luebeck
Intracerebral hemorrhage is one of the diseases with the highest mortality and poorest prognosis worldwide. Spontaneous intracerebral hemorrhage (SICH) typically presents acutely, prompt and expedited radiological examination is crucial for diagnosis, localization, and quantification of the hemorrhage. Early detection and accurate segmentation of perihematomal edema (PHE) play a critical role in guiding appropriate clinical intervention and enhancing patient prognosis. However, the progress and assessment of computer-aided diagnostic methods for PHE segmentation and detection face challenges due to the scarcity of publicly accessible brain CT image datasets. This study establishes a publicly available CT dataset named PHE-SICH-CT-IDS for perihematomal edema in spontaneous intracerebral hemorrhage. The dataset comprises 120 brain CT scans and 7,022 CT images, along with corresponding medical information of the patients. To demonstrate its effectiveness, classical algorithms for semantic segmentation, object detection, and radiomic feature extraction are evaluated. The experimental results confirm the suitability of PHE-SICH-CT-IDS for assessing the performance of segmentation, detection and radiomic feature extraction methods. To the best of our knowledge, this is the first publicly available dataset for PHE in SICH, comprising various data formats suitable for applications across diverse medical scenarios. We believe that PHE-SICH-CT-IDS will allure researchers to explore novel algorithms, providing valuable support for clinicians and patients in the clinical setting. PHE-SICH-CT-IDS is freely published for non-commercial purpose at: this https URL.
In recent years, colorectal cancer has become one of the most significant
diseases that endanger human health. Deep learning methods are increasingly
important for the classification of colorectal histopathology images. However,
existing approaches focus more on end-to-end automatic classification using
computers rather than human-computer interaction. In this paper, we propose an
IL-MCAM framework. It is based on attention mechanisms and interactive
learning. The proposed IL-MCAM framework includes two stages: automatic
learning (AL) and interactivity learning (IL). In the AL stage, a multi-channel
attention mechanism model containing three different attention mechanism
channels and convolutional neural networks is used to extract multi-channel
features for classification. In the IL stage, the proposed IL-MCAM framework
continuously adds misclassified images to the training set in an interactive
approach, which improves the classification ability of the MCAM model. We
carried out a comparison experiment on our dataset and an extended experiment
on the HE-NCT-CRC-100K dataset to verify the performance of the proposed
IL-MCAM framework, achieving classification accuracies of 98.98% and 99.77%,
respectively. In addition, we conducted an ablation experiment and an
interchangeability experiment to verify the ability and interchangeability of
the three channels. The experimental results show that the proposed IL-MCAM
framework has excellent performance in the colorectal histopathological image
classification tasks.
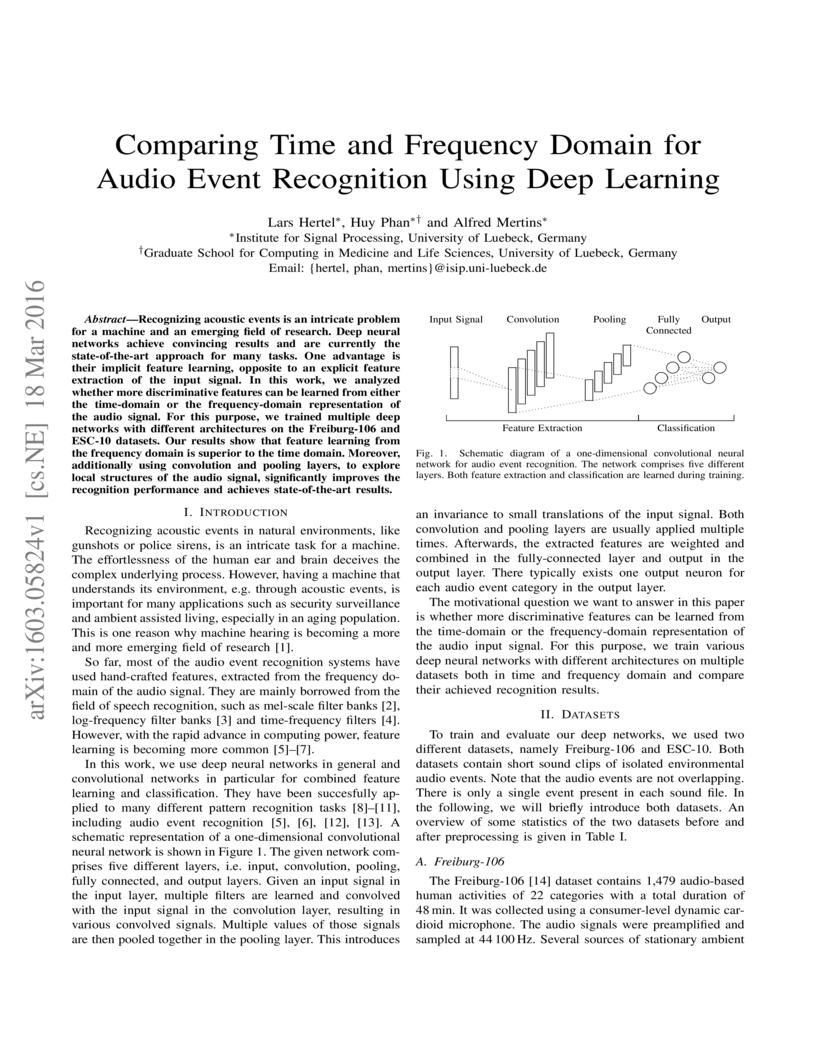
Recognizing acoustic events is an intricate problem for a machine and an emerging field of research. Deep neural networks achieve convincing results and are currently the state-of-the-art approach for many tasks. One advantage is their implicit feature learning, opposite to an explicit feature extraction of the input signal. In this work, we analyzed whether more discriminative features can be learned from either the time-domain or the frequency-domain representation of the audio signal. For this purpose, we trained multiple deep networks with different architectures on the Freiburg-106 and ESC-10 datasets. Our results show that feature learning from the frequency domain is superior to the time domain. Moreover, additionally using convolution and pooling layers, to explore local structures of the audio signal, significantly improves the recognition performance and achieves state-of-the-art results.
Accelerating Magnetic Resonance Imaging (MRI) reduces scan time but often degrades image quality. While Implicit Neural Representations (INRs) show promise for MRI reconstruction, they struggle at high acceleration factors due to weak prior constraints, leading to structural loss and aliasing artefacts. To address this, we propose PrIINeR, an INR-based MRI reconstruction method that integrates prior knowledge from pre-trained deep learning models into the INR framework. By combining population-level knowledge with instance-based optimization and enforcing dual data consistency, PrIINeR aligns both with the acquired k-space data and the prior-informed reconstruction. Evaluated on the NYU fastMRI dataset, our method not only outperforms state-of-the-art INR-based approaches but also improves upon several learning-based state-of-the-art methods, significantly improving structural preservation and fidelity while effectively removing aliasing this http URL bridges deep learning and INR-based techniques, offering a more reliable solution for high-quality, accelerated MRI reconstruction. The code is publicly available on this https URL.
In the past ten years, the computing power of machine vision (MV) has been
continuously improved, and image analysis algorithms have developed rapidly. At
the same time, histopathological slices can be stored as digital images.
Therefore, MV algorithms can provide doctors with diagnostic references. In
particular, the continuous improvement of deep learning algorithms has further
improved the accuracy of MV in disease detection and diagnosis. This paper
reviews the applications of image processing technology based on MV in lymphoma
histopathological images in recent years, including segmentation,
classification and detection. Finally, the current methods are analyzed, some
more potential methods are proposed, and further prospects are made.
Cervical cancer is the seventh most common cancer among all the cancers
worldwide and the fourth most common cancer among women. Cervical cytopathology
image classification is an important method to diagnose cervical cancer. Manual
screening of cytopathology images is time-consuming and error-prone. The
emergence of the automatic computer-aided diagnosis system solves this problem.
This paper proposes a framework called CVM-Cervix based on deep learning to
perform cervical cell classification tasks. It can analyze pap slides quickly
and accurately. CVM-Cervix first proposes a Convolutional Neural Network module
and a Visual Transformer module for local and global feature extraction
respectively, then a Multilayer Perceptron module is designed to fuse the local
and global features for the final classification. Experimental results show the
effectiveness and potential of the proposed CVM-Cervix in the field of cervical
Pap smear image classification. In addition, according to the practical needs
of clinical work, we perform a lightweight post-processing to compress the
model.
The tuning of hyperparameters in distributed machine learning can substantially impact model performance. When the hyperparameters are tuned on sensitive data, privacy becomes an important challenge and to this end, differential privacy has emerged as the de facto standard for provable privacy. A standard setting when performing distributed learning tasks is that clients agree on a shared setup, i.e., find a compromise from a set of hyperparameters, like the learning rate of the model to be trained. Yet, prior work on differentially private hyperparameter tuning either uses computationally expensive cryptographic protocols, determines hyperparameters separately for each client, or applies differential privacy locally, which can lead to undesirable utility-privacy trade-offs.
In this work, we present our algorithm DP-HYPE, which performs a distributed and privacy-preserving hyperparameter search by conducting a distributed voting based on local hyperparameter evaluations of clients. In this way, DP-HYPE selects hyperparameters that lead to a compromise supported by the majority of clients, while maintaining scalability and independence from specific learning tasks. We prove that DP-HYPE preserves the strong notion of differential privacy called client-level differential privacy and, importantly, show that its privacy guarantees do not depend on the number of hyperparameters. We also provide bounds on its utility guarantees, that is, the probability of reaching a compromise, and implement DP-HYPE as a submodule in the popular Flower framework for distributed machine learning. In addition, we evaluate performance on multiple benchmark data sets in iid as well as multiple non-iid settings and demonstrate high utility of DP-HYPE even under small privacy budgets.
Image analysis technology is used to solve the inadvertences of artificial traditional methods in disease, wastewater treatment, environmental change monitoring analysis and convolutional neural networks (CNN) play an important role in microscopic image analysis. An important step in detection, tracking, monitoring, feature extraction, modeling and analysis is image segmentation, in which U-Net has increasingly applied in microscopic image segmentation. This paper comprehensively reviews the development history of U-Net, and analyzes various research results of various segmentation methods since the emergence of U-Net and conducts a comprehensive review of related papers. First, this paper has summarized the improved methods of U-Net and then listed the existing significance of image segmentation techniques and their improvements that has introduced over the years. Finally, focusing on the different improvement strategies of U-Net in different papers, the related work of each application target is reviewed according to detailed technical categories to facilitate future research. Researchers can clearly see the dynamics of transmission of technological development and keep up with future trends in this interdisciplinary field.
30 Dec 2013
Cortical slow oscillations occur in the mammalian brain during deep sleep and
have been shown to contribute to memory consolidation, an effect that can be
enhanced by electrical stimulation. As the precise underlying working
mechanisms are not known it is desired to develop and analyze computational
models of slow oscillations and to study the response to electrical stimuli. In
this paper we employ the conductance based model of Compte et al. [J
Neurophysiol 89, 2707] to study the effect of electrical stimulation. The
population response to electrical stimulation depends on the timing of the
stimulus with respect to the state of the slow oscillation. First, we reproduce
the experimental results of electrical stimulation in ferret brain slices by
Shu et al. [Nature 423, 288] from the conductance based model. We then
numerically obtain the phase response curve for the conductance based network
model to quantify the network's response to weak stimuli. Our results agree
with experiments in vivo and in vitro that show that sensitivity to stimulation
is weaker in the up than in the down state. However, we also find that within
the up state stimulation leads to a shortening of the up state, or phase
advance, whereas during the up-down transition a prolongation of up states is
possible, resulting in a phase delay. Finally, we compute the phase response
curve for the simple mean-field model by Ngo et al. [Europhys Lett 89, 68002]
and find that the qualitative shape of the PRC is preserved, despite its
different mechanism for the generation of slow oscillations.
GasHisSDB: A New Gastric Histopathology Image Dataset for Computer Aided Diagnosis of Gastric Cancer
GasHisSDB: A New Gastric Histopathology Image Dataset for Computer Aided Diagnosis of Gastric Cancer
Background and Objective: Gastric cancer has turned out to be the fifth most
common cancer globally, and early detection of gastric cancer is essential to
save lives. Histopathological examination of gastric cancer is the gold
standard for the diagnosis of gastric cancer. However, computer-aided
diagnostic techniques are challenging to evaluate due to the scarcity of
publicly available gastric histopathology image datasets. Methods: In this
paper, a noble publicly available Gastric Histopathology Sub-size Image
Database (GasHisSDB) is published to identify classifiers' performance.
Specifically, two types of data are included: normal and abnormal, with a total
of 245,196 tissue case images. In order to prove that the methods of different
periods in the field of image classification have discrepancies on GasHisSDB,
we select a variety of classifiers for evaluation. Seven classical machine
learning classifiers, three Convolutional Neural Network classifiers, and a
novel transformer-based classifier are selected for testing on image
classification tasks. Results: This study performed extensive experiments using
traditional machine learning and deep learning methods to prove that the
methods of different periods have discrepancies on GasHisSDB. Traditional
machine learning achieved the best accuracy rate of 86.08% and a minimum of
just 41.12%. The best accuracy of deep learning reached 96.47% and the lowest
was 86.21%. Accuracy rates vary significantly across classifiers. Conclusions:
To the best of our knowledge, it is the first publicly available gastric cancer
histopathology dataset containing a large number of images for weakly
supervised learning. We believe that GasHisSDB can attract researchers to
explore new algorithms for the automated diagnosis of gastric cancer, which can
help physicians and patients in the clinical setting.
This thesis focuses on dealing with the task of acoustic scene classification (ASC), and then applied the techniques developed for ASC to a real-life application of detecting respiratory disease. To deal with ASC challenges, this thesis addresses three main factors that directly affect the performance of an ASC system. Firstly, this thesis explores input features by making use of multiple spectrograms (log-mel, Gamma, and CQT) for low-level feature extraction to tackle the issue of insufficiently discriminative or descriptive input features. Next, a novel Encoder network architecture is introduced. The Encoder firstly transforms each low-level spectrogram into high-level intermediate features, or embeddings, and thus combines these high-level features to form a very distinct composite feature. The composite or combined feature is then explored in terms of classification performance, with different Decoders such as Random Forest (RF), Multilayer Perception (MLP), and Mixture of Experts (MoE). By using this Encoder-Decoder framework, it helps to reduce the computation cost of the reference process in ASC systems which make use of multiple spectrogram inputs. Since the proposed techniques applied for general ASC tasks were shown to be highly effective, this inspired an application to a specific real-life application. This was namely the 2017 Internal Conference on Biomedical Health Informatics (ICBHI) respiratory sound dataset. Building upon the proposed ASC framework, the ICBHI tasks were tackled with a deep learning framework, and the resulting system shown to be capable at detecting respiratory anomaly cycles and diseases.
17 Feb 2022
Background and purpose: Colorectal cancer has become the third most common cancer worldwide, accounting for approximately 10% of cancer patients. Early detection of the disease is important for the treatment of colorectal cancer patients. Histopathological examination is the gold standard for screening colorectal cancer. However, the current lack of histopathological image datasets of colorectal cancer, especially enteroscope biopsies, hinders the accurate evaluation of computer-aided diagnosis techniques. Methods: A new publicly available Enteroscope Biopsy Histopathological H&E Image Dataset (EBHI) is published in this paper. To demonstrate the effectiveness of the EBHI dataset, we have utilized several machine learning, convolutional neural networks and novel transformer-based classifiers for experimentation and evaluation, using an image with a magnification of 200x. Results: Experimental results show that the deep learning method performs well on the EBHI dataset. Traditional machine learning methods achieve maximum accuracy of 76.02% and deep learning method achieves a maximum accuracy of 95.37%. Conclusion: To the best of our knowledge, EBHI is the first publicly available colorectal histopathology enteroscope biopsy dataset with four magnifications and five types of images of tumor differentiation stages, totaling 5532 images. We believe that EBHI could attract researchers to explore new classification algorithms for the automated diagnosis of colorectal cancer, which could help physicians and patients in clinical settings.
15 Apr 2024
In this study, we present a purely data-driven method that uses the Loewner
framework (LF) along with nonlinear optimization techniques to infer quadratic
with affine control dynamical systems that admit Volterra series (VS)
representations from input-output (i/o) time-domain measurements. The proposed
method extensively employs optimization tools for interpolating the symmetric
generalized frequency response functions (GFRFs) derived in the VS framework.
The GFRF estimations are obtained from the Fourier spectrum (phase and
amplitude) of the quasi-steady state system response under harmonic excitation.
Appropriate treatment of these measurements under the developed framework
allows the identification of low-order quadratic state-space models with
non-trivial stable equilibria, such as in the Lorenz '63 forced system. We thus
can achieve low-order global model identification for systems that can
bifurcate to multiple equilibria after solely collecting measurements from a
local stable operational regime. The developed framework is tested for several
examples of increasing dimension and complexity, up to a test case of the
viscous Burgers' equation with Robin boundary conditions. In the latter case,
this study enforces new directions of employing data-driven reduced model
inference that successfully can provide low-order accurate surrogate predictive
models suitable for control. Future directions and open challenges conclude
this work.
Methods: In this study, a benchmark \emph{Abdominal Adipose Tissue CT Image
Dataset} (AATTCT-IDS) containing 300 subjects is prepared and published.
AATTCT-IDS publics 13,732 raw CT slices, and the researchers individually
annotate the subcutaneous and visceral adipose tissue regions of 3,213 of those
slices that have the same slice distance to validate denoising methods, train
semantic segmentation models, and study radiomics. For different tasks, this
paper compares and analyzes the performance of various methods on AATTCT-IDS by
combining the visualization results and evaluation data. Thus, verify the
research potential of this data set in the above three types of tasks.
Results: In the comparative study of image denoising, algorithms using a
smoothing strategy suppress mixed noise at the expense of image details and
obtain better evaluation data. Methods such as BM3D preserve the original image
structure better, although the evaluation data are slightly lower. The results
show significant differences among them. In the comparative study of semantic
segmentation of abdominal adipose tissue, the segmentation results of adipose
tissue by each model show different structural characteristics. Among them,
BiSeNet obtains segmentation results only slightly inferior to U-Net with the
shortest training time and effectively separates small and isolated adipose
tissue. In addition, the radiomics study based on AATTCT-IDS reveals three
adipose distributions in the subject population.
Conclusion: AATTCT-IDS contains the ground truth of adipose tissue regions in
abdominal CT slices. This open-source dataset can attract researchers to
explore the multi-dimensional characteristics of abdominal adipose tissue and
thus help physicians and patients in clinical practice. AATCT-IDS is freely
published for non-commercial purpose at:
\url{this https URL}.
16 Sep 2025
As quantum computing advances, PQC schemes are adopted to replace classical algorithms. Among them is the SLH-DSA that was recently standardized by NIST and is favored for its conservative security foundations.
In this work, we present the first software-only universal forgery attack on SLH-DSA, leveraging Rowhammer-induced bit flips to corrupt the internal state and forge signatures. While prior work targeted embedded systems and required physical access, our attack is software-only, targeting commodity desktop and server hardware, significantly broadening the threat model. We demonstrate a full end-to-end attack against all security levels of SLH-DSA in OpenSSL 3.5.1, achieving universal forgery for the highest security level after eight hours of hammering and 36 seconds of post-processing. Our post-processing is informed by a novel complexity analysis that, given a concrete set of faulty signatures, identifies the most promising computational path to pursue.
To enable the attack, we introduce Swage, a modular and extensible framework for implementing end-to-end Rowhammer-based fault attacks. Swage abstracts and automates key components of practical Rowhammer attacks. Unlike prior tooling, Swage is untangled from the attacked code, making it reusable and suitable for frictionless analysis of different targets. Our findings highlight that even theoretically sound PQC schemes can fail under real-world conditions, underscoring the need for additional implementation hardening or hardware defenses against Rowhammer.
The gold standard for gastric cancer detection is gastric histopathological
image analysis, but there are certain drawbacks in the existing
histopathological detection and diagnosis. In this paper, based on the study of
computer aided diagnosis system, graph based features are applied to gastric
cancer histopathology microscopic image analysis, and a classifier is used to
classify gastric cancer cells from benign cells. Firstly, image segmentation is
performed, and after finding the region, cell nuclei are extracted using the
k-means method, the minimum spanning tree (MST) is drawn, and graph based
features of the MST are extracted. The graph based features are then put into
the classifier for classification. In this study, different segmentation
methods are compared in the tissue segmentation stage, among which are
Level-Set, Otsu thresholding, watershed, SegNet, U-Net and Trans-U-Net
segmentation; Graph based features, Red, Green, Blue features, Grey-Level
Co-occurrence Matrix features, Histograms of Oriented Gradient features and
Local Binary Patterns features are compared in the feature extraction stage;
Radial Basis Function (RBF) Support Vector Machine (SVM), Linear SVM,
Artificial Neural Network, Random Forests, k-NearestNeighbor, VGG16, and
Inception-V3 are compared in the classifier stage. It is found that using U-Net
to segment tissue areas, then extracting graph based features, and finally
using RBF SVM classifier gives the optimal results with 94.29%.
With the acceleration of urbanization and living standards, microorganisms
play increasingly important roles in industrial production, bio-technique, and
food safety testing. Microorganism biovolume measurements are one of the
essential parts of microbial analysis. However, traditional manual measurement
methods are time-consuming and challenging to measure the characteristics
precisely. With the development of digital image processing techniques, the
characteristics of the microbial population can be detected and quantified. The
changing trend can be adjusted in time and provided a basis for the
improvement. The applications of the microorganism biovolume measurement method
have developed since the 1980s. More than 62 articles are reviewed in this
study, and the articles are grouped by digital image segmentation methods with
periods. This study has high research significance and application value, which
can be referred to microbial researchers to have a comprehensive understanding
of microorganism biovolume measurements using digital image analysis methods
and potential applications.
24 Aug 2021
Controlling large-scale particle or robot systems is challenging because of
their high dimensionality. We use a centralized stochastic approach that allows
for optimal control at the cost of a central element instead of a decentralized
approach. Previous works are often restricted to the assumption of fully
actuated robots. Here we propose an approach for underactuated robots that
allows for energy-efficient control of the robot system. We consider a simple
task of gathering the robots (minimizing positional variance) and steering them
towards a goal point within a bounded area without obstacles. We make two main
contributions. First, we present a generalized coordinate transformation for
underactuated robots, whose physical properties should be considered. We choose
Euler- Lagrange systems that describe a large class of robot systems. Second,
we propose an optimal control mechanism with the prime objective of energy
efficiency. We show the feasibility of our approach in numerical simulations
and robot simulations.
Current virtual reality (VR) training simulators of liver punctures often rely on static 3D patient data and use an unrealistic (sinusoidal) periodic animation of the respiratory movement. Existing methods for the animation of breathing motion support simple mathematical or patient-specific, estimated breathing models. However with personalized breathing models for each new patient, a heavily dose relevant or expensive 4D data acquisition is mandatory for keyframe-based motion modeling. Given the reference 4D data, first a model building stage using linear regression motion field modeling takes place. Then the methodology shown here allows the transfer of existing reference respiratory motion models of a 4D reference patient to a new static 3D patient. This goal is achieved by using non-linear inter-patient registration to warp one personalized 4D motion field model to new 3D patient data. This cost- and dose-saving new method is shown here visually in a qualitative proof-of-concept study.
12 Apr 2018
Research on automated vehicles has experienced an explosive growth over the past decade. A main obstacle to their practical realization, however, is a convincing safety concept. This question becomes ever more important as more sophisticated algorithms are used and the vehicle automation level increases. The field of functional safety offers a systematic approach to identify possible sources of risk and to improve the safety of a vehicle. It is based on practical experience across the aerospace, process and other industries over multiple decades. This experience is compiled in the functional safety standard for the automotive domain, ISO 26262, which is widely adopted throughout the automotive industry. However, its applicability and relevance for highly automated vehicles is subject to a controversial debate. This paper takes a critical look at the discussion and summarizes the main steps of ISO 26262 for a safe control design for automated vehicles.
There are no more papers matching your filters at the moment.