University of Massachusetts Medical School
02 Jun 2025
This work introduces RARE (Retrieval-Augmented Reasoning Enhancement), a
versatile extension to the mutual reasoning framework (rStar), aimed at
enhancing reasoning accuracy and factual integrity across large language models
(LLMs) for complex, knowledge-intensive tasks such as commonsense and medical
reasoning. RARE incorporates two innovative actions within the Monte Carlo Tree
Search (MCTS) framework: A6, which generates search queries based on the
initial problem statement, performs information retrieval using those queries,
and augments reasoning with the retrieved data to formulate the final answer;
and A7, which leverages information retrieval specifically for generated
sub-questions and re-answers these sub-questions with the relevant contextual
information. Additionally, a Retrieval-Augmented Factuality Scorer is proposed
to replace the original discriminator, prioritizing reasoning paths that meet
high standards of factuality. Experimental results with LLaMA 3.1 show that
RARE enables open-source LLMs to achieve competitive performance with top
open-source models like GPT-4 and GPT-4o. This research establishes RARE as a
scalable solution for improving LLMs in domains where logical coherence and
factual integrity are critical.
To enhance the performance of large language models (LLMs) in biomedical natural language processing (BioNLP) by introducing a domain-specific instruction dataset and examining its impact when combined with multi-task learning principles. We created the BioInstruct, comprising 25,005 instructions to instruction-tune LLMs(LLaMA 1 & 2, 7B & 13B version). The instructions were created by prompting the GPT-4 language model with three-seed samples randomly drawn from an 80 human curated instructions. We employed Low-Rank Adaptation(LoRA) for parameter-efficient fine-tuning. We then evaluated these instruction-tuned LLMs on several BioNLP tasks, which can be grouped into three major categories: question answering(QA), information extraction(IE), and text generation(GEN). We also examined whether categories(e.g., QA, IE, and generation) of instructions impact model performance. Comparing with LLMs without instruction-tuned, our instruction-tuned LLMs demonstrated marked performance gains: 17.3% in QA, 5.7% in IE, and 96% in Generation tasks. Our 7B-parameter instruction-tuned LLaMA 1 model was competitive or even surpassed other LLMs in the biomedical domain that were also fine-tuned from LLaMA 1 with vast domain-specific data or a variety of tasks. Our results also show that the performance gain is significantly higher when instruction fine-tuning is conducted with closely related tasks. Our findings align with the observations of multi-task learning, suggesting the synergies between two tasks. The BioInstruct dataset serves as a valuable resource and instruction tuned LLMs lead to the best performing BioNLP applications.
Large Language Models (LLMs) have demonstrated a remarkable potential in medical knowledge acquisition and question-answering. However, LLMs can potentially hallucinate and yield factually incorrect outcomes, even with domain-specific pretraining. Previously, retrieval augmented generation (RAG) has limited success in addressing hallucinations. Unlike previous methods in RAG where the retrieval model was trained separately from the LLM, we introduce JMLR (for Jointly trains LLM and information Retrieval) during the fine-tuning phase. The synchronized training mechanism enhances JMLR's ability to retrieve clinical guidelines and leverage medical knowledge to reason and answer questions and reduces the demand for computational resources. We evaluated JMLR on the important medical question-answering application. Our experimental results demonstrate that JMLR-13B (70.5%) outperforms a previous state-of-the-art open-source model using conventional pre-training and fine-tuning Meditron-70B (68.9%) and Llama2-13B with RAG (67.7%) on a medical question-answering dataset. Comprehensive evaluations reveal JMLR-13B enhances reasoning quality and reduces hallucinations better than Claude3-Opus. Additionally, JMLR-13B (148 GPU hours) also trains much faster than Meditron-70B (42630 GPU hours). Through this work, we provide a new and efficient knowledge enhancement method for healthcare, demonstrating the potential of integrating retrieval and LLM training for medical question-answering systems.
Multimodal large language models (MLLMs) have made significant strides, yet they face challenges in the medical domain due to limited specialized knowledge. While recent medical MLLMs demonstrate strong performance in lab settings, they often struggle in real-world applications, highlighting a substantial gap between research and practice. In this paper, we seek to address this gap at various stages of the end-to-end learning pipeline, including data collection, model fine-tuning, and evaluation. At the data collection stage, we introduce SemiHVision, a dataset that combines human annotations with automated augmentation techniques to improve both medical knowledge representation and diagnostic reasoning. For model fine-tuning, we trained PMC-Cambrian-8B-AN over 2400 H100 GPU hours, resulting in performance that surpasses public medical models like HuatuoGPT-Vision-34B (79.0% vs. 66.7%) and private general models like Claude3-Opus (55.7%) on traditional benchmarks such as SLAKE and VQA-RAD. In the evaluation phase, we observed that traditional benchmarks cannot accurately reflect realistic clinical task capabilities. To overcome this limitation and provide more targeted guidance for model evaluation, we introduce the JAMA Clinical Challenge, a novel benchmark specifically designed to evaluate diagnostic reasoning. On this benchmark, PMC-Cambrian-AN achieves state-of-the-art performance with a GPT-4 score of 1.29, significantly outperforming HuatuoGPT-Vision-34B (1.13) and Claude3-Opus (1.17), demonstrating its superior diagnostic reasoning abilities.
Generative AI has demonstrated strong potential in healthcare, from clinical decision support to patient-facing chatbots that improve outcomes. A critical challenge for deployment is effective human-AI communication, where content must be both personalized and understandable. We introduce MedReadCtrl, a readability-controlled instruction tuning framework that enables LLMs to adjust output complexity without compromising meaning. Evaluations of nine datasets and three tasks across medical and general domains show that MedReadCtrl achieves significantly lower readability instruction-following errors than GPT-4 (e.g., 1.39 vs. 1.59 on ReadMe, p<0.001) and delivers substantial gains on unseen clinical tasks (e.g., +14.7 ROUGE-L, +6.18 SARI on MTSamples). Experts consistently preferred MedReadCtrl (71.7% vs. 23.3%), especially at low literacy levels. These gains reflect MedReadCtrl's ability to restructure clinical content into accessible, readability-aligned language while preserving medical intent, offering a scalable solution to support patient education and expand equitable access to AI-enabled care.
Translating electronic health record (EHR) narratives from English to Spanish is a clinically important yet challenging task due to the lack of a parallel-aligned corpus and the abundant unknown words contained. To address such challenges, we propose \textbf{NOOV} (for No OOV), a new neural machine translation (NMT) system that requires little in-domain parallel-aligned corpus for training. NOOV integrates a bilingual lexicon automatically learned from parallel-aligned corpora and a phrase look-up table extracted from a large biomedical knowledge resource, to alleviate both the unknown word problem and the word-repeat challenge in NMT, enhancing better phrase generation of NMT systems. Evaluation shows that NOOV is able to generate better translation of EHR with improvement in both accuracy and fluency.
All cognitive agents are composite beings. Specifically, complex living
agents consist of cells, which are themselves competent sub-agents navigating
physiological and metabolic spaces. Behavior science, evolutionary
developmental biology, and the field of machine intelligence all seek an answer
to the scaling of biological cognition: what evolutionary dynamics enable
individual cells to integrate their activities to result in the emergence of a
novel, higher-level intelligence that has goals and competencies that belong to
it and not to its parts? Here, we report the results of simulations based on
the TAME framework, which proposes that evolution pivoted the collective
intelligence of cells during morphogenesis of the body into traditional
behavioral intelligence by scaling up the goal states at the center of
homeostatic processes. We tested the hypothesis that a minimal evolutionary
framework is sufficient for small, low-level setpoints of metabolic homeostasis
in cells to scale up into collectives (tissues) which solve a problem in
morphospace: the organization of a body-wide positional information axis (the
classic French Flag problem). We found that these emergent morphogenetic agents
exhibit a number of predicted features, including the use of stress propagation
dynamics to achieve its target morphology as well as the ability to recover
from perturbation (robustness) and long-term stability (even though neither of
these was directly selected for). Moreover we observed unexpected behavior of
sudden remodeling long after the system stabilizes. We tested this prediction
in a biological system - regenerating planaria - and observed a very similar
phenomenon. We propose that this system is a first step toward a quantitative
understanding of how evolution scales minimal goal-directed behavior
(homeostatic loops) into higher-level problem-solving agents in morphogenetic
and other spaces.
Eviction is a significant yet understudied social determinants of health (SDoH), linked to housing instability, unemployment, and mental health. While eviction appears in unstructured electronic health records (EHRs), it is rarely coded in structured fields, limiting downstream applications. We introduce SynthEHR-Eviction, a scalable pipeline combining LLMs, human-in-the-loop annotation, and automated prompt optimization (APO) to extract eviction statuses from clinical notes. Using this pipeline, we created the largest public eviction-related SDoH dataset to date, comprising 14 fine-grained categories. Fine-tuned LLMs (e.g., Qwen2.5, LLaMA3) trained on SynthEHR-Eviction achieved Macro-F1 scores of 88.8% (eviction) and 90.3% (other SDoH) on human validated data, outperforming GPT-4o-APO (87.8%, 87.3%), GPT-4o-mini-APO (69.1%, 78.1%), and BioBERT (60.7%, 68.3%), while enabling cost-effective deployment across various model sizes. The pipeline reduces annotation effort by over 80%, accelerates dataset creation, enables scalable eviction detection, and generalizes to other information extraction tasks.
20 May 2023
Objective: Evictions are important social and behavioral determinants of
health. Evictions are associated with a cascade of negative events that can
lead to unemployment, housing insecurity/homelessness, long-term poverty, and
mental health problems. In this study, we developed a natural language
processing system to automatically detect eviction status from electronic
health record (EHR) notes.
Materials and Methods: We first defined eviction status (eviction presence
and eviction period) and then annotated eviction status in 5000 EHR notes from
the Veterans Health Administration (VHA). We developed a novel model, KIRESH,
that has shown to substantially outperform other state-of-the-art models such
as fine-tuning pre-trained language models like BioBERT and BioClinicalBERT.
Moreover, we designed a novel prompt to further improve the model performance
by using the intrinsic connection between the two sub-tasks of eviction
presence and period prediction. Finally, we used the Temperature Scaling-based
Calibration on our KIRESH-Prompt method to avoid over-confidence issues arising
from the imbalance dataset.
Results: KIRESH-Prompt substantially outperformed strong baseline models
including fine-tuning the BioClinicalBERT model to achieve 0.74672 MCC, 0.71153
Macro-F1, and 0.83396 Micro-F1 in predicting eviction period and 0.66827 MCC,
0.62734 Macro-F1, and 0.7863 Micro-F1 in predicting eviction presence. We also
conducted additional experiments on a benchmark social determinants of health
(SBDH) dataset to demonstrate the generalizability of our methods.
Conclusion and Future Work: KIRESH-Prompt has substantially improved eviction
status classification. We plan to deploy KIRESH-Prompt to the VHA EHRs as an
eviction surveillance system to help address the US Veterans' housing
insecurity.
26 Sep 2017
With the recent focus in the accessibility field, researchers from academia
and industry have been very active in developing innovative techniques and
tools for assistive technology. Especially with handheld devices getting ever
powerful and being able to recognize the user's voice, screen magnification for
individuals with low-vision, and eye tracking devices used in studies with
individuals with physical and intellectual disabilities, the science field is
quickly adapting and creating conclusions as well as products to help. In this
paper, we will focus on new technology and tools to help make reading
easier--including reformatting document presentation (for people with physical
vision impairments) and text simplification to make information itself easier
to interpret (for people with intellectual disabilities). A real-world case
study is reported based on our experience to make documents more accessible.
Efficient and accurate detection of subtle motion generated from small
objects in noisy environments, as needed for vital sign monitoring, is
challenging, but can be substantially improved with magnification. We developed
a complex Gabor filter-based decomposition method to amplify phases at
different spatial wavelength levels to magnify motion and extract 1D motion
signals for fundamental frequency estimation. The phase-based complex Gabor
filter outputs are processed and then used to train machine learning models
that predict respiration and heart rate with greater accuracy. We show that our
proposed technique performs better than the conventional temporal FFT-based
method in clinical settings, such as sleep laboratories and emergency
departments, as well for a variety of human postures.
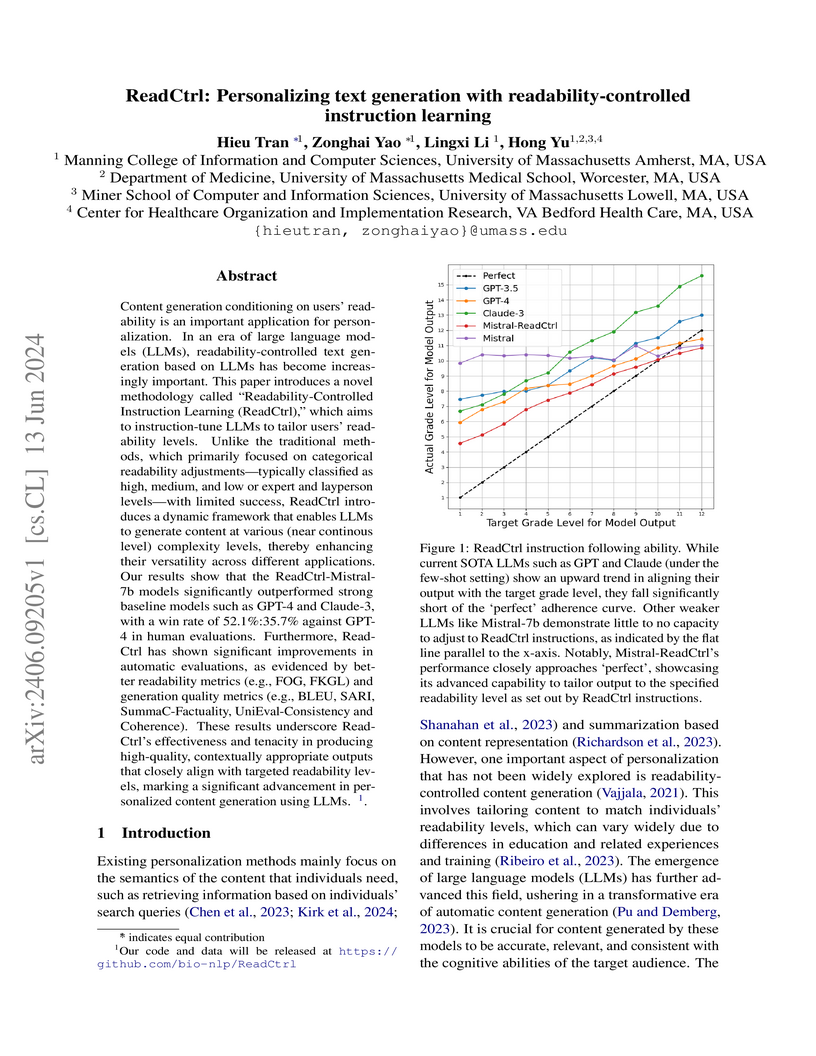
ReadCtrl fine-tunes language models to generate text at specific readability levels, allowing for personalized content creation across a spectrum of reading abilities. The system, developed by researchers at the University of Massachusetts, demonstrated superior readability control compared to GPT-4 and Claude-3, achieving a 52.1% win rate against GPT-4 in human evaluations.
Wikipedia (Wiki) is one of the most widely used and publicly available resources for natural language processing (NLP) applications. Wikipedia Revision History (WikiRevHist) shows the order in which edits were made to any Wiki page since its first modification. While the most up-to-date Wiki has been widely used as a training source, WikiRevHist can also be valuable resources for NLP applications. However, there are insufficient tools available to process WikiRevHist without having substantial computing resources, making additional customization, and spending extra time adapting others' works. Therefore, we report Blocks Architecture (BloArk), an efficiency-focused data processing architecture that reduces running time, computing resource requirements, and repeated works in processing WikiRevHist dataset. BloArk consists of three parts in its infrastructure: blocks, segments, and warehouses. On top of that, we build the core data processing pipeline: builder and modifier. The BloArk builder transforms the original WikiRevHist dataset from XML syntax into JSON Lines (JSONL) format for improving the concurrent and storage efficiency. The BloArk modifier takes previously-built warehouses to operate incremental modifications for improving the utilization of existing databases and reducing the cost of reusing others' works. In the end, BloArk can scale up easily in both processing Wikipedia Revision History and incrementally modifying existing dataset for downstream NLP use cases. The source code, documentations, and example usages are publicly available online and open-sourced under GPL-2.0 license.
Language Models (LMs) have performed well on biomedical natural language processing applications. In this study, we conducted some experiments to use prompt methods to extract knowledge from LMs as new knowledge Bases (LMs as KBs). However, prompting can only be used as a low bound for knowledge extraction, and perform particularly poorly on biomedical domain KBs. In order to make LMs as KBs more in line with the actual application scenarios of the biomedical domain, we specifically add EHR notes as context to the prompt to improve the low bound in the biomedical domain. We design and validate a series of experiments for our Dynamic-Context-BioLAMA task. Our experiments show that the knowledge possessed by those language models can distinguish the correct knowledge from the noise knowledge in the EHR notes, and such distinguishing ability can also be used as a new metric to evaluate the amount of knowledge possessed by the model.
03 Sep 2025
Real-world adoption of closed-loop insulin delivery systems (CLIDS) in type 1 diabetes remains low, driven not by technical failure, but by diverse behavioral, psychosocial, and social barriers. We introduce ChatCLIDS, the first benchmark to rigorously evaluate LLM-driven persuasive dialogue for health behavior change. Our framework features a library of expert-validated virtual patients, each with clinically grounded, heterogeneous profiles and realistic adoption barriers, and simulates multi-turn interactions with nurse agents equipped with a diverse set of evidence-based persuasive strategies. ChatCLIDS uniquely supports longitudinal counseling and adversarial social influence scenarios, enabling robust, multi-dimensional evaluation. Our findings reveal that while larger and more reflective LLMs adapt strategies over time, all models struggle to overcome resistance, especially under realistic social pressure. These results highlight critical limitations of current LLMs for behavior change, and offer a high-fidelity, scalable testbed for advancing trustworthy persuasive AI in healthcare and beyond.
Objective: Allowing patients to access their own electronic health record
(EHR) notes through online patient portals has the potential to improve
patient-centered care. However, medical jargon, which abounds in EHR notes, has
been shown to be a barrier for patient EHR comprehension. Existing knowledge
bases that link medical jargon to lay terms or definitions play an important
role in alleviating this problem but have low coverage of medical jargon in
EHRs. We developed a data-driven approach that mines EHRs to identify and rank
medical jargon based on its importance to patients, to support the building of
EHR-centric lay language resources.
Methods: We developed an innovative adapted distant supervision (ADS) model
based on support vector machines to rank medical jargon from EHRs. For distant
supervision, we utilized the open-access, collaborative consumer health
vocabulary, a large, publicly available resource that links lay terms to
medical jargon. We explored both knowledge-based features from the Unified
Medical Language System and distributed word representations learned from
unlabeled large corpora. We evaluated the ADS model using physician-identified
important medical terms.
Results: Our ADS model significantly surpassed two state-of-the-art automatic
term recognition methods, TF*IDF and C-Value, yielding 0.810 ROC-AUC versus
0.710 and 0.667, respectively. Our model identified 10K important medical
jargon terms after ranking over 100K candidate terms mined from over 7,500 EHR
narratives.
Conclusion: Our work is an important step towards enriching lexical resources
that link medical jargon to lay terms/definitions to support patient EHR
comprehension. The identified medical jargon terms and their rankings are
available upon request.
Coherence plays a critical role in producing a high-quality summary from a
document. In recent years, neural extractive summarization is becoming
increasingly attractive. However, most of them ignore the coherence of
summaries when extracting sentences. As an effort towards extracting coherent
summaries, we propose a neural coherence model to capture the cross-sentence
semantic and syntactic coherence patterns. The proposed neural coherence model
obviates the need for feature engineering and can be trained in an end-to-end
fashion using unlabeled data. Empirical results show that the proposed neural
coherence model can efficiently capture the cross-sentence coherence patterns.
Using the combined output of the neural coherence model and ROUGE package as
the reward, we design a reinforcement learning method to train a proposed
neural extractive summarizer which is named Reinforced Neural Extractive
Summarization (RNES) model. The RNES model learns to optimize coherence and
informative importance of the summary simultaneously. Experimental results show
that the proposed RNES outperforms existing baselines and achieves
state-of-the-art performance in term of ROUGE on CNN/Daily Mail dataset. The
qualitative evaluation indicates that summaries produced by RNES are more
coherent and readable.
 CNRS
CNRS University of Oxford
University of Oxford McGill University
McGill University KU LeuvenRadboud UniversityTU DresdenNational Institute of Standards and TechnologyInstitut CurieEcole Polytechnique Fédérale de LausanneFederal Institute for Materials Research and TestingINSERM
KU LeuvenRadboud UniversityTU DresdenNational Institute of Standards and TechnologyInstitut CurieEcole Polytechnique Fédérale de LausanneFederal Institute for Materials Research and TestingINSERM University of BaselUniversity of GothenburgNewcastle UniversityPSL Research UniversityHoward Hughes Medical InstituteUniversity of DundeeThe Rockefeller UniversityEuropean Molecular Biology LaboratoryUniversity of Massachusetts Medical SchoolFriedrich Miescher Institute for Biomedical ResearchResearch Institute of Molecular PathologyVIBEuro-BioImagingMax Planck Institute for Heart and Lung ResearchCancer Research UK Manchester InstituteTurku Bioscience CentreMax Planck Institute of Immunobiology and EpigeneticsMax Planck Institute of Experimental MedicineMax Planck Institute for Biology of Ageing
University of BaselUniversity of GothenburgNewcastle UniversityPSL Research UniversityHoward Hughes Medical InstituteUniversity of DundeeThe Rockefeller UniversityEuropean Molecular Biology LaboratoryUniversity of Massachusetts Medical SchoolFriedrich Miescher Institute for Biomedical ResearchResearch Institute of Molecular PathologyVIBEuro-BioImagingMax Planck Institute for Heart and Lung ResearchCancer Research UK Manchester InstituteTurku Bioscience CentreMax Planck Institute of Immunobiology and EpigeneticsMax Planck Institute of Experimental MedicineMax Planck Institute for Biology of AgeingIn April 2020, the QUality Assessment and REProducibility for Instruments and
Images in Light Microscopy (QUAREP-LiMi) initiative was formed. This initiative
comprises imaging scientists from academia and industry who share a common
interest in achieving a better understanding of the performance and limitations
of microscopes and improved quality control (QC) in light microscopy. The
ultimate goal of the QUAREP-LiMi initiative is to establish a set of common QC
standards, guidelines, metadata models, and tools, including detailed
protocols, with the ultimate aim of improving reproducible advances in
scientific research. This White Paper 1) summarizes the major obstacles
identified in the field that motivated the launch of the QUAREP-LiMi
initiative; 2) identifies the urgent need to address these obstacles in a
grassroots manner, through a community of stakeholders including, researchers,
imaging scientists, bioimage analysts, bioimage informatics developers,
corporate partners, funding agencies, standards organizations, scientific
publishers, and observers of such; 3) outlines the current actions of the
QUAREP-LiMi initiative, and 4) proposes future steps that can be taken to
improve the dissemination and acceptance of the proposed guidelines to manage
QC. To summarize, the principal goal of the QUAREP-LiMi initiative is to
improve the overall quality and reproducibility of light microscope image data
by introducing broadly accepted standard practices and accurately captured
image data metrics.
Antigen-specific T cells play an essential role in immunoregulation and
diseases such as cancer. Characterizing the T cell receptor (TCR) sequences
that encode T cell specificity is critical for elucidating the antigenic
determinants of immunological diseases and designing therapeutic remedies.
However, methods of obtaining single-cell TCR sequencing data are labor and
cost intensive, requiring cell sorting and full length single-cell
RNA-sequencing (scRNA-seq). New high-throughput 3' cell-barcoding scRNA-seq
methods can simplify and scale this process; however, they do not routinely
capture TCR sequences during library preparation and sequencing. While 5'
cell-barcoding scRNA-seq methods can be used to examine TCR repertoire at
single-cell resolution, it requires specialized reagents which cannot be
applied to samples previously processed using 3' cell-barcoding methods. Here,
we outline a method for sequencing TCRα and TCRβ transcripts from
samples already processed using 3' cell-barcoding scRNA-seq platforms, ensuring
TCR recovery at a single-cell resolution. In short, a fraction of the 3'
barcoded whole transcriptome amplification (WTA) product typically used to
generate a massively parallel 3' scRNA-seq library is enriched for TCR
transcripts using biotinylated probes, and further amplified using the same
universal primer sequence from WTA. Primer extension using TCR V-region primers
and targeted PCR amplification results in a 3' barcoded single-cell
CDR3-enriched library that can be sequenced with custom sequencing primers.
Coupled with 3' scRNA-seq of the same WTA, this method enables simultaneous
analysis of single-cell transcriptomes and TCR sequences which can help
interpret inherent heterogeneity among antigen-specific T cells and salient
disease biology. This method can be adapted to enrich other transcripts of
interest from 3' and 5' barcoded WTA libraries.
University of Zurich University College LondonTechnische Universitat Munchen
University College LondonTechnische Universitat Munchen Shanghai Jiao Tong University
Shanghai Jiao Tong University Zhejiang University
Zhejiang University University of Copenhagen
University of Copenhagen The Chinese University of Hong Kong
The Chinese University of Hong Kong King’s College London
King’s College London Technical University of MunichErasmus MCUniversity Medical Center UtrechtUniversity of Massachusetts Medical SchoolHarbin Institute of Technology at ShenzhenEPITAPasqual Maragall FoundationAmsterdam University Medical CentreElectronique de ParisH. Lundbeck A/SHospital del Mar Medical Research Institute (IMIM)ISEP-Institut Sup ́erieur d’GlaxoSmithKline Research":
Technical University of MunichErasmus MCUniversity Medical Center UtrechtUniversity of Massachusetts Medical SchoolHarbin Institute of Technology at ShenzhenEPITAPasqual Maragall FoundationAmsterdam University Medical CentreElectronique de ParisH. Lundbeck A/SHospital del Mar Medical Research Institute (IMIM)ISEP-Institut Sup ́erieur d’GlaxoSmithKline Research":
 University College LondonTechnische Universitat Munchen
University College LondonTechnische Universitat Munchen Shanghai Jiao Tong University
Shanghai Jiao Tong University Zhejiang University
Zhejiang University University of Copenhagen
University of Copenhagen The Chinese University of Hong Kong
The Chinese University of Hong Kong King’s College London
King’s College London Technical University of MunichErasmus MCUniversity Medical Center UtrechtUniversity of Massachusetts Medical SchoolHarbin Institute of Technology at ShenzhenEPITAPasqual Maragall FoundationAmsterdam University Medical CentreElectronique de ParisH. Lundbeck A/SHospital del Mar Medical Research Institute (IMIM)ISEP-Institut Sup ́erieur d’GlaxoSmithKline Research":
Technical University of MunichErasmus MCUniversity Medical Center UtrechtUniversity of Massachusetts Medical SchoolHarbin Institute of Technology at ShenzhenEPITAPasqual Maragall FoundationAmsterdam University Medical CentreElectronique de ParisH. Lundbeck A/SHospital del Mar Medical Research Institute (IMIM)ISEP-Institut Sup ́erieur d’GlaxoSmithKline Research":Imaging markers of cerebral small vessel disease provide valuable information on brain health, but their manual assessment is time-consuming and hampered by substantial intra- and interrater variability. Automated rating may benefit biomedical research, as well as clinical assessment, but diagnostic reliability of existing algorithms is unknown. Here, we present the results of the \textit{VAscular Lesions DetectiOn and Segmentation} (\textit{Where is VALDO?}) challenge that was run as a satellite event at the international conference on Medical Image Computing and Computer Aided Intervention (MICCAI) 2021. This challenge aimed to promote the development of methods for automated detection and segmentation of small and sparse imaging markers of cerebral small vessel disease, namely enlarged perivascular spaces (EPVS) (Task 1), cerebral microbleeds (Task 2) and lacunes of presumed vascular origin (Task 3) while leveraging weak and noisy labels. Overall, 12 teams participated in the challenge proposing solutions for one or more tasks (4 for Task 1 - EPVS, 9 for Task 2 - Microbleeds and 6 for Task 3 - Lacunes). Multi-cohort data was used in both training and evaluation. Results showed a large variability in performance both across teams and across tasks, with promising results notably for Task 1 - EPVS and Task 2 - Microbleeds and not practically useful results yet for Task 3 - Lacunes. It also highlighted the performance inconsistency across cases that may deter use at an individual level, while still proving useful at a population level.
There are no more papers matching your filters at the moment.