Ping An Technology
This paper provides a comprehensive survey of safety considerations in Large Language Models (LLMs)
Researchers from Tianjin University and Ping An Technology introduced ChatSOP, a framework that leverages Standard Operating Procedures (SOPs) and Monte Carlo Tree Search (MCTS) to guide Large Language Model dialogue agents. This approach yields substantial improvements in dialogue controllability, proactivity, and overall task success compared to existing methods.
07 Sep 2025
Researchers from Shandong University, Ludwig Maximilian University of Munich, Ping An Technology, and Computility Lab developed MV-Debate, a multi-view agent debate framework with dynamic reflection gating for multimodal harmful content detection in social media. This framework employs specialized agents to analyze explicit, implicit, cross-modal, and contextual nuances, achieving new state-of-the-art results with an average accuracy of 84.9% and F1 score of 78.0% across sarcasm, hate speech, and misinformation detection tasks.
The field of deep generative modeling is dominated by generative adversarial
networks (GANs). However, the training of GANs often lacks stability, fails to
converge, and suffers from model collapse. It takes an assortment of tricks to
solve these problems, which may be difficult to understand for those seeking to
apply generative modeling. Instead, we propose two novel generative
autoencoders, AE-OTtrans and AE-OTgen, which rely on optimal transport instead
of adversarial training. AE-OTtrans and AEOTgen, unlike VAE and WAE, preserve
the manifold of the data; they do not force the latent distribution to match a
normal distribution, resulting in greater quality images. AEOTtrans and
AE-OTgen also produce images of higher diversity compared to their predecessor,
AE-OT. We show that AE-OTtrans and AE-OTgen surpass GANs in the MNIST and
FashionMNIST datasets. Furthermore, We show that AE-OTtrans and AE-OTgen do
state of the art on the MNIST, FashionMNIST, and CelebA image sets comapred to
other non-adversarial generative models.
Image landmark detection aims to automatically identify the locations of
predefined fiducial points. Despite recent success in this field,
higher-ordered structural modeling to capture implicit or explicit
relationships among anatomical landmarks has not been adequately exploited. In
this work, we present a new topology-adapting deep graph learning approach for
accurate anatomical facial and medical (e.g., hand, pelvis) landmark detection.
The proposed method constructs graph signals leveraging both local image
features and global shape features. The adaptive graph topology naturally
explores and lands on task-specific structures which are learned end-to-end
with two Graph Convolutional Networks (GCNs). Extensive experiments are
conducted on three public facial image datasets (WFLW, 300W, and COFW-68) as
well as three real-world X-ray medical datasets (Cephalometric (public), Hand
and Pelvis). Quantitative results comparing with the previous state-of-the-art
approaches across all studied datasets indicating the superior performance in
both robustness and accuracy. Qualitative visualizations of the learned graph
topologies demonstrate a physically plausible connectivity laying behind the
landmarks.
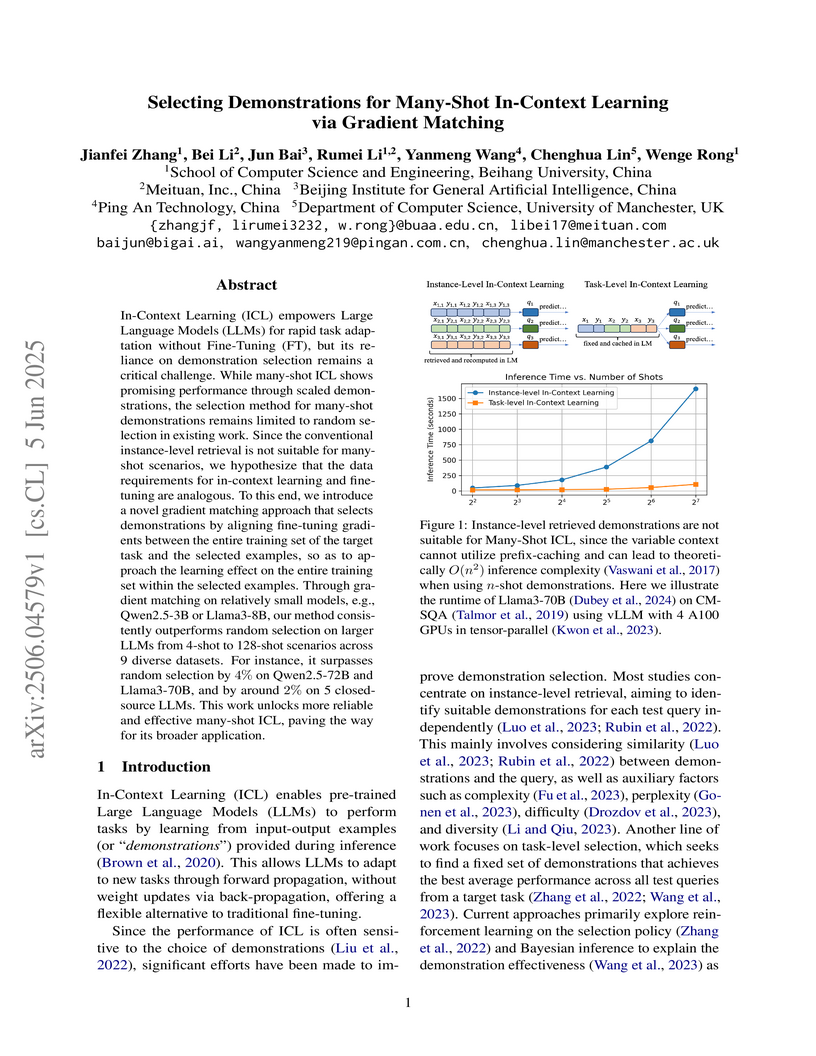
In-Context Learning (ICL) empowers Large Language Models (LLMs) for rapid
task adaptation without Fine-Tuning (FT), but its reliance on demonstration
selection remains a critical challenge. While many-shot ICL shows promising
performance through scaled demonstrations, the selection method for many-shot
demonstrations remains limited to random selection in existing work. Since the
conventional instance-level retrieval is not suitable for many-shot scenarios,
we hypothesize that the data requirements for in-context learning and
fine-tuning are analogous. To this end, we introduce a novel gradient matching
approach that selects demonstrations by aligning fine-tuning gradients between
the entire training set of the target task and the selected examples, so as to
approach the learning effect on the entire training set within the selected
examples. Through gradient matching on relatively small models, e.g.,
Qwen2.5-3B or Llama3-8B, our method consistently outperforms random selection
on larger LLMs from 4-shot to 128-shot scenarios across 9 diverse datasets. For
instance, it surpasses random selection by 4% on Qwen2.5-72B and Llama3-70B,
and by around 2% on 5 closed-source LLMs. This work unlocks more reliable and
effective many-shot ICL, paving the way for its broader application.
Most earlier researches on talking face generation have focused on the
synchronization of lip motion and speech content. However, head pose and facial
emotions are equally important characteristics of natural faces. While
audio-driven talking face generation has seen notable advancements, existing
methods either overlook facial emotions or are limited to specific individuals
and cannot be applied to arbitrary subjects. In this paper, we propose a novel
one-shot Talking Head Generation framework (SPEAK) that distinguishes itself
from the general Talking Face Generation by enabling emotional and postural
control. Specifically, we introduce Inter-Reconstructed Feature Disentanglement
(IRFD) module to decouple facial features into three latent spaces. Then we
design a face editing module that modifies speech content and facial latent
codes into a single latent space. Subsequently, we present a novel generator
that employs modified latent codes derived from the editing module to regulate
emotional expression, head poses, and speech content in synthesizing facial
animations. Extensive trials demonstrate that our method ensures lip
synchronization with the audio while enabling decoupled control of facial
features, it can generate realistic talking head with coordinated lip motions,
authentic facial emotions, and smooth head movements. The demo video is
available: this https URL
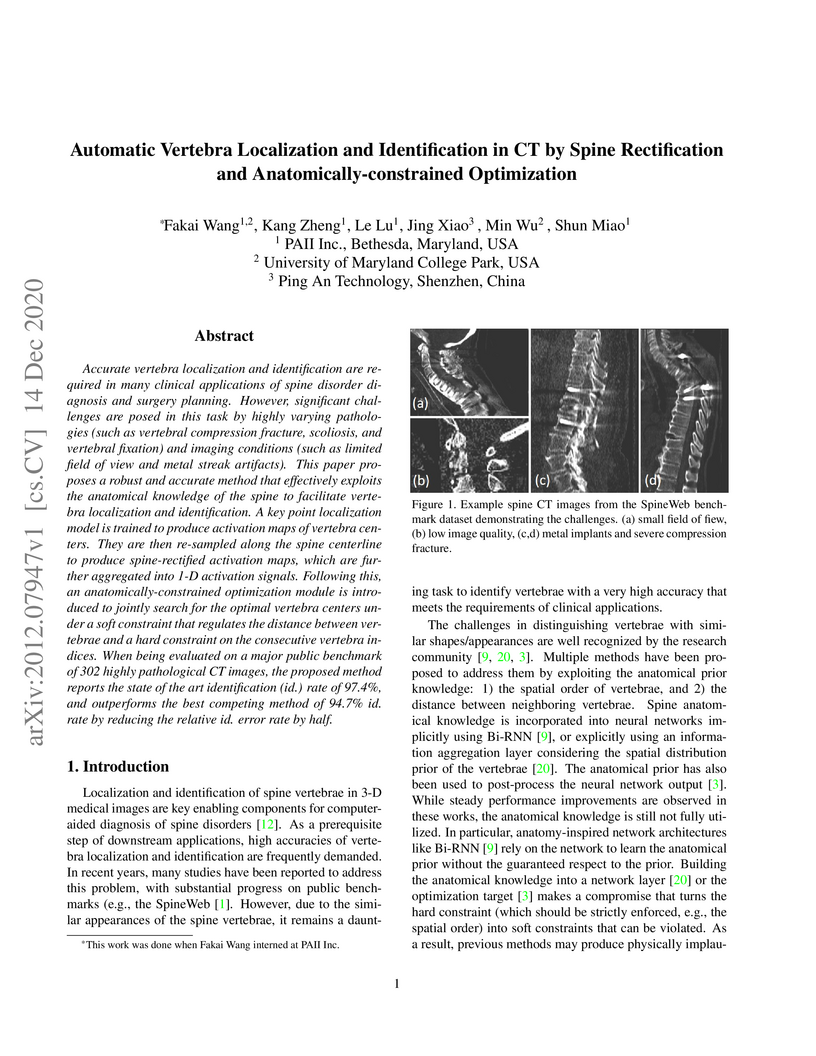
Accurate vertebra localization and identification are required in many
clinical applications of spine disorder diagnosis and surgery planning.
However, significant challenges are posed in this task by highly varying
pathologies (such as vertebral compression fracture, scoliosis, and vertebral
fixation) and imaging conditions (such as limited field of view and metal
streak artifacts). This paper proposes a robust and accurate method that
effectively exploits the anatomical knowledge of the spine to facilitate
vertebra localization and identification. A key point localization model is
trained to produce activation maps of vertebra centers. They are then
re-sampled along the spine centerline to produce spine-rectified activation
maps, which are further aggregated into 1-D activation signals. Following this,
an anatomically-constrained optimization module is introduced to jointly search
for the optimal vertebra centers under a soft constraint that regulates the
distance between vertebrae and a hard constraint on the consecutive vertebra
indices. When being evaluated on a major public benchmark of 302 highly
pathological CT images, the proposed method reports the state of the art
identification (id.) rate of 97.4%, and outperforms the best competing method
of 94.7% id. rate by reducing the relative id. error rate by half.
Visual cues of enforcing bilaterally symmetric anatomies as normal findings are widely used in clinical practice to disambiguate subtle abnormalities from medical images. So far, inadequate research attention has been received on effectively emulating this practice in CAD methods. In this work, we exploit semantic anatomical symmetry or asymmetry analysis in a complex CAD scenario, i.e., anterior pelvic fracture detection in trauma PXRs, where semantically pathological (refer to as fracture) and non-pathological (e.g., pose) asymmetries both occur. Visually subtle yet pathologically critical fracture sites can be missed even by experienced clinicians, when limited diagnosis time is permitted in emergency care. We propose a novel fracture detection framework that builds upon a Siamese network enhanced with a spatial transformer layer to holistically analyze symmetric image features. Image features are spatially formatted to encode bilaterally symmetric anatomies. A new contrastive feature learning component in our Siamese network is designed to optimize the deep image features being more salient corresponding to the underlying semantic asymmetries (caused by pelvic fracture occurrences). Our proposed method have been extensively evaluated on 2,359 PXRs from unique patients (the largest study to-date), and report an area under ROC curve score of 0.9771. This is the highest among state-of-the-art fracture detection methods, with improved clinical indications.
The rocketing prosperity of large language models (LLMs) in recent years has boosted the prevalence of vision-language models (VLMs) in the medical sector. In our online medical consultation scenario, a doctor responds to the texts and images provided by a patient in multiple rounds to diagnose her/his health condition, forming a multi-turn multimodal medical dialogue format. Unlike high-quality images captured by professional equipment in traditional medical visual question answering (Med-VQA), the images in our case are taken by patients' mobile phones. These images have poor quality control, with issues such as excessive background elements and the lesion area being significantly off-center, leading to degradation of vision-language alignment in the model training phase. In this paper, we propose ZALM3, a Zero-shot strategy to improve vision-language ALignment in Multi-turn Multimodal Medical dialogue. Since we observe that the preceding text conversations before an image can infer the regions of interest (RoIs) in the image, ZALM3 employs an LLM to summarize the keywords from the preceding context and a visual grounding model to extract the RoIs. The updated images eliminate unnecessary background noise and provide more effective vision-language alignment. To better evaluate our proposed method, we design a new subjective assessment metric for multi-turn unimodal/multimodal medical dialogue to provide a fine-grained performance comparison. Our experiments across three different clinical departments remarkably demonstrate the efficacy of ZALM3 with statistical significance.
The pancreatic disease taxonomy includes ten types of masses (tumors or
cysts)[20,8]. Previous work focuses on developing segmentation or
classification methods only for certain mass types. Differential diagnosis of
all mass types is clinically highly desirable [20] but has not been
investigated using an automated image understanding approach. We exploit the
feasibility to distinguish pancreatic ductal adenocarcinoma (PDAC) from the
nine other nonPDAC masses using multi-phase CT imaging. Both image appearance
and the 3D organ-mass geometry relationship are critical. We propose a holistic
segmentation-mesh-classification network (SMCN) to provide patient-level
diagnosis, by fully utilizing the geometry and location information, which is
accomplished by combining the anatomical structure and the semantic
detection-by-segmentation network. SMCN learns the pancreas and mass
segmentation task and builds an anatomical correspondence-aware organ mesh
model by progressively deforming a pancreas prototype on the raw segmentation
mask (i.e., mask-to-mesh). A new graph-based residual convolutional network
(Graph-ResNet), whose nodes fuse the information of the mesh model and feature
vectors extracted from the segmentation network, is developed to produce the
patient-level differential classification results. Extensive experiments on 661
patients' CT scans (five phases per patient) show that SMCN can improve the
mass segmentation and detection accuracy compared to the strong baseline method
nnUNet (e.g., for nonPDAC, Dice: 0.611 vs. 0.478; detection rate: 89% vs. 70%),
achieve similar sensitivity and specificity in differentiating PDAC and nonPDAC
as expert radiologists (i.e., 94% and 90%), and obtain results comparable to a
multimodality test [20] that combines clinical, imaging, and molecular testing
for clinical management of patients.
Hip and pelvic fractures are serious injuries with life-threatening
complications. However, diagnostic errors of fractures in pelvic X-rays (PXRs)
are very common, driving the demand for computer-aided diagnosis (CAD)
solutions. A major challenge lies in the fact that fractures are localized
patterns that require localized analyses. Unfortunately, the PXRs residing in
hospital picture archiving and communication system do not typically specify
region of interests. In this paper, we propose a two-stage hip and pelvic
fracture detection method that executes localized fracture classification using
weakly supervised ROI mining. The first stage uses a large capacity
fully-convolutional network, i.e., deep with high levels of abstraction, in a
multiple instance learning setting to automatically mine probable true positive
and definite hard negative ROIs from the whole PXR in the training data. The
second stage trains a smaller capacity model, i.e., shallower and more
generalizable, with the mined ROIs to perform localized analyses to classify
fractures. During inference, our method detects hip and pelvic fractures in one
pass by chaining the probability outputs of the two stages together. We
evaluate our method on 4 410 PXRs, reporting an area under the ROC curve value
of 0.975, the highest among state-of-the-art fracture detection methods.
Moreover, we show that our two-stage approach can perform comparably to human
physicians (even outperforming emergency physicians and surgeons), in a
preliminary reader study of 23 readers.
In medical imaging, organ/pathology segmentation models trained on current publicly available and fully-annotated datasets usually do not well-represent the heterogeneous modalities, phases, pathologies, and clinical scenarios encountered in real environments. On the other hand, there are tremendous amounts of unlabelled patient imaging scans stored by many modern clinical centers. In this work, we present a novel segmentation strategy, co-heterogenous and adaptive segmentation (CHASe), which only requires a small labeled cohort of single phase imaging data to adapt to any unlabeled cohort of heterogenous multi-phase data with possibly new clinical scenarios and pathologies. To do this, we propose a versatile framework that fuses appearance based semi-supervision, mask based adversarial domain adaptation, and pseudo-labeling. We also introduce co-heterogeneous training, which is a novel integration of co-training and hetero modality learning. We have evaluated CHASe using a clinically comprehensive and challenging dataset of multi-phase computed tomography (CT) imaging studies (1147 patients and 4577 3D volumes). Compared to previous state-of-the-art baselines, CHASe can further improve pathological liver mask Dice-Sorensen coefficients by ranges of 4.2%∼9.4%, depending on the phase combinations: e.g., from 84.6% to 94.0% on non-contrast CTs.
Bone mineral density (BMD) is a clinically critical indicator of osteoporosis, usually measured by dual-energy X-ray absorptiometry (DEXA). Due to the limited accessibility of DEXA machines and examinations, osteoporosis is often under-diagnosed and under-treated, leading to increased fragility fracture risks. Thus it is highly desirable to obtain BMDs with alternative cost-effective and more accessible medical imaging examinations such as X-ray plain films. In this work, we formulate the BMD estimation from plain hip X-ray images as a regression problem. Specifically, we propose a new semi-supervised self-training algorithm to train the BMD regression model using images coupled with DEXA measured BMDs and unlabeled images with pseudo BMDs. Pseudo BMDs are generated and refined iteratively for unlabeled images during self-training. We also present a novel adaptive triplet loss to improve the model's regression accuracy. On an in-house dataset of 1,090 images (819 unique patients), our BMD estimation method achieves a high Pearson correlation coefficient of 0.8805 to ground-truth BMDs. It offers good feasibility to use the more accessible and cheaper X-ray imaging for opportunistic osteoporosis screening.
How to produce expressive molecular representations is a fundamental
challenge in AI-driven drug discovery. Graph neural network (GNN) has emerged
as a powerful technique for modeling molecular data. However, previous
supervised approaches usually suffer from the scarcity of labeled data and have
poor generalization capability. Here, we proposed a novel Molecular
Pre-training Graph-based deep learning framework, named MPG, that leans
molecular representations from large-scale unlabeled molecules. In MPG, we
proposed a powerful MolGNet model and an effective self-supervised strategy for
pre-training the model at both the node and graph-level. After pre-training on
11 million unlabeled molecules, we revealed that MolGNet can capture valuable
chemistry insights to produce interpretable representation. The pre-trained
MolGNet can be fine-tuned with just one additional output layer to create
state-of-the-art models for a wide range of drug discovery tasks, including
molecular properties prediction, drug-drug interaction, and drug-target
interaction, involving 13 benchmark datasets. Our work demonstrates that MPG is
promising to become a novel approach in the drug discovery pipeline.
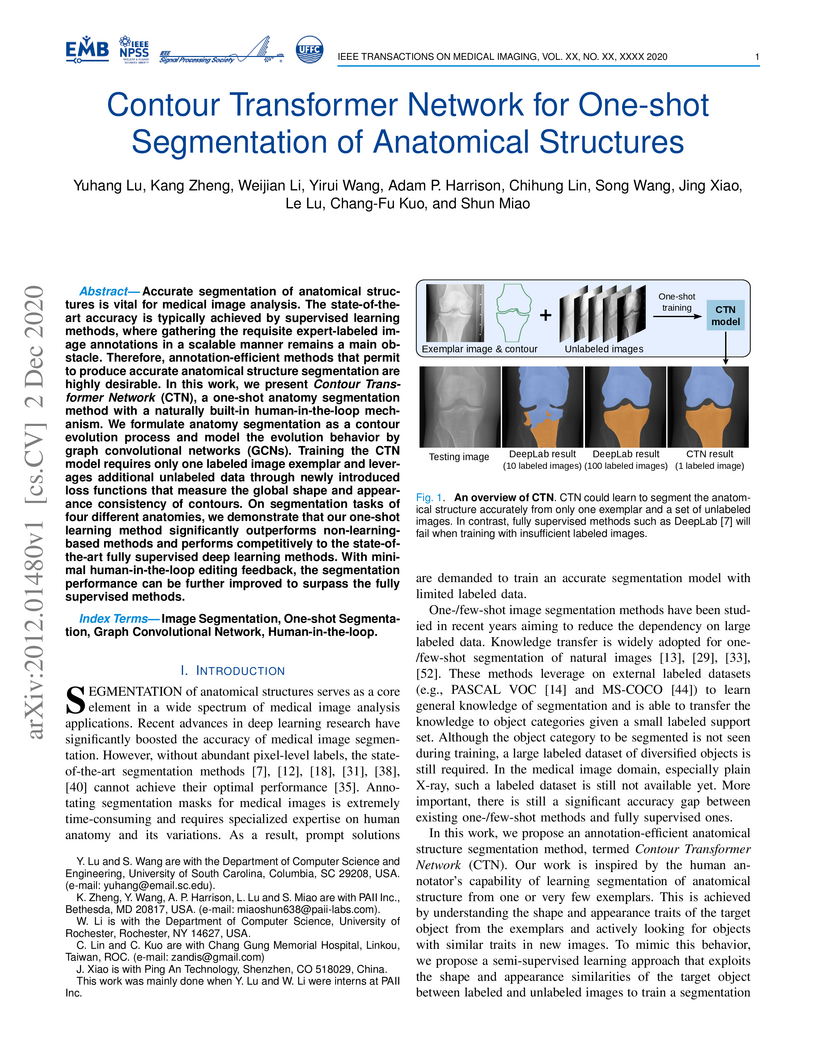
Accurate segmentation of anatomical structures is vital for medical image analysis. The state-of-the-art accuracy is typically achieved by supervised learning methods, where gathering the requisite expert-labeled image annotations in a scalable manner remains a main obstacle. Therefore, annotation-efficient methods that permit to produce accurate anatomical structure segmentation are highly desirable. In this work, we present Contour Transformer Network (CTN), a one-shot anatomy segmentation method with a naturally built-in human-in-the-loop mechanism. We formulate anatomy segmentation as a contour evolution process and model the evolution behavior by graph convolutional networks (GCNs). Training the CTN model requires only one labeled image exemplar and leverages additional unlabeled data through newly introduced loss functions that measure the global shape and appearance consistency of contours. On segmentation tasks of four different anatomies, we demonstrate that our one-shot learning method significantly outperforms non-learning-based methods and performs competitively to the state-of-the-art fully supervised deep learning methods. With minimal human-in-the-loop editing feedback, the segmentation performance can be further improved to surpass the fully supervised methods.
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal cancers and
carries a dismal prognosis. Surgery remains the best chance of a potential cure
for patients who are eligible for initial resection of PDAC. However, outcomes
vary significantly even among the resected patients of the same stage and
received similar treatments. Accurate preoperative prognosis of resectable
PDACs for personalized treatment is thus highly desired. Nevertheless, there
are no automated methods yet to fully exploit the contrast-enhanced computed
tomography (CE-CT) imaging for PDAC. Tumor attenuation changes across different
CT phases can reflect the tumor internal stromal fractions and vascularization
of individual tumors that may impact the clinical outcomes. In this work, we
propose a novel deep neural network for the survival prediction of resectable
PDAC patients, named as 3D Contrast-Enhanced Convolutional Long Short-Term
Memory network(CE-ConvLSTM), which can derive the tumor attenuation signatures
or patterns from CE-CT imaging studies. We present a multi-task CNN to
accomplish both tasks of outcome and margin prediction where the network
benefits from learning the tumor resection margin related features to improve
survival prediction. The proposed framework can improve the prediction
performances compared with existing state-of-the-art survival analysis
approaches. The tumor signature built from our model has evidently added values
to be combined with the existing clinical staging system.
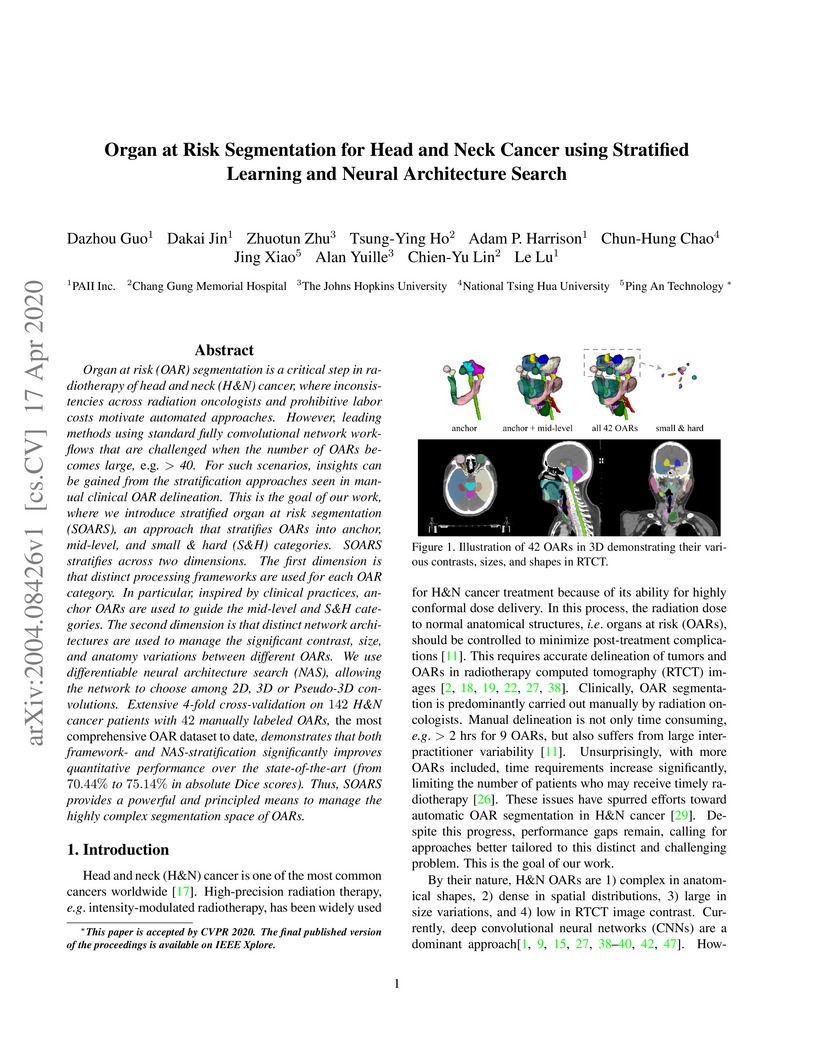
OAR segmentation is a critical step in radiotherapy of head and neck (H&N)
cancer, where inconsistencies across radiation oncologists and prohibitive
labor costs motivate automated approaches. However, leading methods using
standard fully convolutional network workflows that are challenged when the
number of OARs becomes large, e.g. > 40. For such scenarios, insights can be
gained from the stratification approaches seen in manual clinical OAR
delineation. This is the goal of our work, where we introduce stratified organ
at risk segmentation (SOARS), an approach that stratifies OARs into anchor,
mid-level, and small & hard (S&H) categories. SOARS stratifies across two
dimensions. The first dimension is that distinct processing pipelines are used
for each OAR category. In particular, inspired by clinical practices, anchor
OARs are used to guide the mid-level and S&H categories. The second dimension
is that distinct network architectures are used to manage the significant
contrast, size, and anatomy variations between different OARs. We use
differentiable neural architecture search (NAS), allowing the network to choose
among 2D, 3D or Pseudo-3D convolutions. Extensive 4-fold cross-validation on
142 H&N cancer patients with 42 manually labeled OARs, the most comprehensive
OAR dataset to date, demonstrates that both pipeline- and NAS-stratification
significantly improves quantitative performance over the state-of-the-art (from
69.52% to 73.68% in absolute Dice scores). Thus, SOARS provides a powerful and
principled means to manage the highly complex segmentation space of OARs.
The animated Graphical Interchange Format (GIF) images have been widely used
on social media as an intuitive way of expression emotion. Given their
expressiveness, GIFs offer a more nuanced and precise way to convey emotions.
In this paper, we present our solution for the EmotionGIF 2020 challenge, the
shared task of SocialNLP 2020. To recommend GIF categories for unlabeled
tweets, we regarded this problem as a kind of matching tasks and proposed a
learning to rank framework based on Bidirectional Encoder Representations from
Transformer (BERT) and LightGBM. Our team won the 4th place with a Mean Average
Precision @ 6 (MAP@6) score of 0.5394 on the round 1 leaderboard.
22 Mar 2020
One-Shot methods have evolved into one of the most popular methods in Neural
Architecture Search (NAS) due to weight sharing and single training of a
supernet. However, existing methods generally suffer from two issues:
predetermined number of channels in each layer which is suboptimal; and model
averaging effects and poor ranking correlation caused by weight coupling and
continuously expanding search space. To explicitly address these issues, in
this paper, a Broadening-and-Shrinking One-Shot NAS (BS-NAS) framework is
proposed, in which `broadening' refers to broadening the search space with a
spring block enabling search for numbers of channels during training of the
supernet; while `shrinking' refers to a novel shrinking strategy gradually
turning off those underperforming operations. The above innovations broaden the
search space for wider representation and then shrink it by gradually removing
underperforming operations, followed by an evolutionary algorithm to
efficiently search for the optimal architecture. Extensive experiments on
ImageNet illustrate the effectiveness of the proposed BS-NAS as well as the
state-of-the-art performance.
There are no more papers matching your filters at the moment.