University of Nebraska Medical Center
05 Sep 2025
 Harvard University
Harvard University University of Manchester
University of Manchester University of Oxford
University of Oxford University of California, Irvine
University of California, Irvine Stanford University
Stanford University Columbia UniversityHarvard Medical School
Columbia UniversityHarvard Medical School Rice UniversityThe University of SydneyDeakin UniversityHarvard T.H. Chan School of Public HealthStellenbosch UniversityBaylor College of MedicineUniversity of Colorado Anschutz Medical CampusUniversity of Nebraska Medical CenterCentre for Addiction and Mental HealthCharité—Universitätsmedizin BerlinPontificia Universidad JaverianaLaureate Institute for Brain ResearchThe University of Texas Southwestern Medical CenterNational Institute of Mental Health and NeurosciencesBrandenburg Medical School Theodor FontaneNational Institute for Health and Care Research (NIHR) Oxford Health Biomedical Research CentreImmanuel Hospital RüdersdorfNational Alliance on Mental Illness (NAMI)Beth-Israel Deaconess Medical Center
Rice UniversityThe University of SydneyDeakin UniversityHarvard T.H. Chan School of Public HealthStellenbosch UniversityBaylor College of MedicineUniversity of Colorado Anschutz Medical CampusUniversity of Nebraska Medical CenterCentre for Addiction and Mental HealthCharité—Universitätsmedizin BerlinPontificia Universidad JaverianaLaureate Institute for Brain ResearchThe University of Texas Southwestern Medical CenterNational Institute of Mental Health and NeurosciencesBrandenburg Medical School Theodor FontaneNational Institute for Health and Care Research (NIHR) Oxford Health Biomedical Research CentreImmanuel Hospital RüdersdorfNational Alliance on Mental Illness (NAMI)Beth-Israel Deaconess Medical CenterIndividuals are increasingly utilizing large language model (LLM)based tools for mental health guidance and crisis support in place of human experts. While AI technology has great potential to improve health outcomes, insufficient empirical evidence exists to suggest that AI technology can be deployed as a clinical replacement; thus, there is an urgent need to assess and regulate such tools. Regulatory efforts have been made and multiple evaluation frameworks have been proposed, however,field-wide assessment metrics have yet to be formally integrated. In this paper, we introduce a comprehensive online platform that aggregates evaluation approaches and serves as a dynamic online resource to simplify LLM and LLM-based tool assessment: MindBenchAI. At its core, MindBenchAI is designed to provide easily accessible/interpretable information for diverse stakeholders (patients, clinicians, developers, regulators, etc.). To create MindBenchAI, we built off our work developing this http URL to support informed decision-making around smartphone app use for mental health, and expanded the technical this http URL framework to encompass novel large language model (LLM) functionalities through benchmarking approaches. The MindBenchAI platform is designed as a partnership with the National Alliance on Mental Illness (NAMI) to provide assessment tools that systematically evaluate LLMs and LLM-based tools with objective and transparent criteria from a healthcare standpoint, assessing both profile (i.e. technical features, privacy protections, and conversational style) and performance characteristics (i.e. clinical reasoning skills).
12 Nov 2024
Medical imaging has played a pivotal role in advancing and refining digital twin technology, allowing for the development of highly personalized virtual models that represent human anatomy and physiological functions. A key component in constructing these digital twins is the integration of high-resolution imaging data, such as MRI, CT, PET, and ultrasound, with sophisticated computational models. Advances in medical imaging significantly enhance real-time simulation, predictive modeling, and early disease diagnosis, individualized treatment planning, ultimately boosting precision and personalized care. Although challenges persist, such as the complexity of anatomical modeling, integrating various imaging modalities, and high computational demands, recent progress in imaging and machine learning has greatly improved the precision and clinical applicability of digital twins. This review investigates the role of medical imaging in developing digital twins across organ systems. Key findings demonstrate that improvements in medical imaging have enhanced the diagnostic and therapeutic potential of digital twins beyond traditional methods, particularly in imaging accuracy, treatment effectiveness, and patient outcomes. The review also examines the technical barriers that currently limit further development of digital twin technology, despite advances in medical imaging, and outlines future research avenues aimed at overcoming these challenges to unlock the full potential of this technology in precision medicine.
15 Sep 2025
There is a challenge in selecting high-dimensional mediators when the mediators have complex correlation structures and interactions. In this work, we frame the high-dimensional mediator selection problem into a series of hypothesis tests with composite nulls, and develop a method to control the false discovery rate (FDR) which has mild assumptions on the mediation model. We show the theoretical guarantee that the proposed method and algorithm achieve FDR control. We present extensive simulation results to demonstrate the power and finite sample performance compared with existing methods. Lastly, we demonstrate the method for analyzing the Alzheimer's Disease Neuroimaging Initiative (ADNI) data, in which the proposed method selects the volume of the hippocampus and amygdala, as well as some other important MRI-derived measures as mediators for the relationship between gender and dementia progression.
Climate change is threatening human health in unprecedented orders and many
ways. These threats are expected to grow unless effective and evidence-based
policies are developed and acted upon to minimize or eliminate them. Attaining
such a task requires the highest degree of the flow of knowledge from science
into policy. The multidisciplinary, location-specific, and vastness of
published science makes it challenging to keep track of novel work in this
area, as well as making the traditional knowledge synthesis methods inefficient
in infusing science into policy. To this end, we consider developing multiple
domain-specific language models (LMs) with different variations from Climate-
and Health-related information, which can serve as a foundational step toward
capturing available knowledge to enable solving different tasks, such as
detecting similarities between climate- and health-related concepts,
fact-checking, relation extraction, evidence of health effects to policy text
generation, and more. To our knowledge, this is the first work that proposes
developing multiple domain-specific language models for the considered domains.
We will make the developed models, resources, and codebase available for the
researchers.
16 Apr 2025
Osteoarthritis (OA) is a prevalent degenerative joint disease, with the knee
being the most commonly affected joint. Modern studies of knee joint injury and
OA often measure biomechanical variables, particularly forces exerted during
walking. However, the relationship among gait patterns, clinical profiles, and
OA disease remains poorly understood. These biomechanical forces are typically
represented as curves over time, but until recently, studies have relied on
discrete values (or landmarks) to summarize these curves. This work aims to
demonstrate the added value of analyzing full movement curves over conventional
discrete summaries. Using data from the Intensive Diet and Exercise for
Arthritis (IDEA) study (Messier et al., 2009, 2013), we developed a shape-based
representation of variation in the full biomechanical curves. Compared to
conventional discrete summaries, our approach yields more powerful predictors
of disease severity and relevant clinical traits, as demonstrated by a nested
model comparison. Notably, our work is among the first to use movement curves
to predict disease measures and to quantitatively evaluate the added value of
analyzing full movement curves over conventional discrete summaries.
23 Jun 2025
This paper introduces a tamper-resistant framework for large language models (LLMs) in medical applications, utilizing quantum gradient descent (QGD) to detect malicious parameter modifications in real time. Integrated into a LLaMA-based model, QGD monitors weight amplitude distributions, identifying adversarial fine-tuning anomalies. Tests on the MIMIC and eICU datasets show minimal performance impact (accuracy: 89.1 to 88.3 on MIMIC) while robustly detecting tampering. PubMedQA evaluations confirm preserved biomedical question-answering capabilities. Compared to baselines like selective unlearning and cryptographic fingerprinting, QGD offers superior sensitivity to subtle weight changes. This quantum-inspired approach ensures secure, reliable medical AI, extensible to other high-stakes domains.
05 Sep 2022
Diabetes is a major public health challenge worldwide. Abnormal physiology in diabetes, particularly hypoglycemia, can cause driver impairments that affect safe driving. While diabetes driver safety has been previously researched, few studies link real-time physiologic changes in drivers with diabetes to objective real-world driver safety, particularly at high-risk areas like intersections. To address this, we investigated the role of acute physiologic changes in drivers with type 1 diabetes mellitus (T1DM) on safe stopping at stop intersections. 18 T1DM drivers (21-52 years, mean = 31.2 years) and 14 controls (21-55 years, mean = 33.4 years) participated in a 4-week naturalistic driving study. At induction, each participant's vehicle was fitted with a camera and sensor system to collect driving data. Video was processed with computer vision algorithms detecting traffic elements. Stop intersections were geolocated with clustering methods, state intersection databases, and manual review. Videos showing driver stop intersection approaches were extracted and manually reviewed to classify stopping behavior (full, rolling, and no stop) and intersection traffic characteristics. Mixed-effects logistic regression models determined how diabetes driver stopping safety (safe vs. unsafe stop) was affected by 1) disease and 2) at-risk, acute physiology (hypo- and hyperglycemia). Diabetes drivers who were acutely hyperglycemic had 2.37 increased odds of unsafe stopping (95% CI: 1.26-4.47, p = 0.008) compared to those with normal physiology. Acute hypoglycemia did not associate with unsafe stopping (p = 0.537), however the lower frequency of hypoglycemia (vs. hyperglycemia) warrants a larger sample of drivers to investigate this effect. Critically, presence of diabetes alone did not associate with unsafe stopping, underscoring the need to evaluate driver physiology in licensing guidelines.
30 Oct 2018
To explain individual differences in development, behavior, and cognition, most previous studies focused on projecting resting-state functional MRI (fMRI) based functional connectivity (FC) data into a low-dimensional space via linear dimensionality reduction techniques, followed by executing analysis operations. However, linear dimensionality analysis techniques may fail to capture nonlinearity of brain neuroactivity. Moreover, besides resting-state FC, FC based on task fMRI can be expected to provide complementary information. Motivated by these considerations, we nonlinearly fuse resting-state and task-based FC networks (FCNs) to seek a better representation in this paper. We propose a framework based on alternating diffusion map (ADM), which extracts geometry-preserving low-dimensional embeddings that successfully parameterize the intrinsic variables driving the phenomenon of interest. Specifically, we first separately build resting-state and task-based FCNs by symmetric positive definite matrices using sparse inverse covariance estimation for each subject, and then utilize the ADM to fuse them in order to extract significant low-dimensional embeddings, which are used as fingerprints to identify individuals. The proposed framework is validated on the Philadelphia Neurodevelopmental Cohort data, where we conduct extensive experimental study on resting-state and fractal n-back task fMRI for the classification of intelligence quotient (IQ). The fusion of resting-state and n-back task fMRI by the proposed framework achieves better classification accuracy than any single fMRI, and the proposed framework is shown to outperform several other data fusion methods. To our knowledge, this paper is the first to demonstrate a successful extension of the ADM to fuse resting-state and task-based fMRI data for accurate prediction of IQ.
Objective: Longitudinal neuroimaging studies have demonstrated that
adolescence is the crucial developmental epoch of continued brain growth and
change. A large number of researchers dedicate to uncovering the mechanisms
about brain maturity during adolescence. Motivated by both achievement in graph
signal processing and recent evidence that some brain areas act as hubs
connecting functionally specialized systems, we proposed an approach to detect
these regions from spectral analysis perspective. In particular, as human brain
undergoes substantial development throughout adolescence, we addressed the
challenge by evaluating the functional network difference among age groups from
functional magnetic resonance imaging (fMRI) observations. Methods: We treated
these observations as graph signals defined on the parcellated functional brain
regions and applied graph Laplacian learning based Fourier Transform (GLFT) to
transform the original graph signals into frequency domain. Eigen-analysis was
conducted afterwards to study the behavior of the corresponding brain regions,
which enables the characterization of brain maturation. Result: We first
evaluated our method on the synthetic data and further applied the method to
resting and task state fMRI imaging data from Philadelphia Neurodevelopmental
Cohort (PNC) dataset, comprised of normally developing adolescents from 8 to
22. The model provided a highest accuracy of 95.69% in distinguishing different
adolescence stages. Conclusion: We detected 13 hubs from resting state fMRI and
16 hubs from task state fMRI that are highly related to brain maturation
process. Significance: The proposed GLFT method is powerful in extracting the
brain connectivity patterns and identifying hub regions with a high prediction
power
Pancreatic ductal adenocarcinoma (PDAC) presents a critical global health challenge, and early detection is crucial for improving the 5-year survival rate. Recent medical imaging and computational algorithm advances offer potential solutions for early diagnosis. Deep learning, particularly in the form of convolutional neural networks (CNNs), has demonstrated success in medical image analysis tasks, including classification and segmentation. However, the limited availability of clinical data for training purposes continues to provide a significant obstacle. Data augmentation, generative adversarial networks (GANs), and cross-validation are potential techniques to address this limitation and improve model performance, but effective solutions are still rare for 3D PDAC, where contrast is especially poor owing to the high heterogeneity in both tumor and background tissues. In this study, we developed a new GAN-based model, named 3DGAUnet, for generating realistic 3D CT images of PDAC tumors and pancreatic tissue, which can generate the interslice connection data that the existing 2D CT image synthesis models lack. Our innovation is to develop a 3D U-Net architecture for the generator to improve shape and texture learning for PDAC tumors and pancreatic tissue. Our approach offers a promising path to tackle the urgent requirement for creative and synergistic methods to combat PDAC. The development of this GAN-based model has the potential to alleviate data scarcity issues, elevate the quality of synthesized data, and thereby facilitate the progression of deep learning models to enhance the accuracy and early detection of PDAC tumors, which could profoundly impact patient outcomes. Furthermore, this model has the potential to be adapted to other types of solid tumors, hence making significant contributions to the field of medical imaging in terms of image processing models.
21 Jan 2025
In this study, we present the Physical-Bio Translator, an agent-based simulation model designed to simulate cellular responses following irradiation. This simulation framework is based on a novel cell-state transition model that accurately reflects the characteristics of irradiated cells. To validate the Physical-Bio Translator, we performed simulations of cell phase evolution, cell phenotype evolution, and cell survival. The results indicate that the Physical-Bio Translator effectively replicates experimental cell irradiation outcomes, suggesting that digital cell irradiation experiments can be conducted via computer simulation, offering a more sophisticated model for radiation biology. This work lays the foundation for developing a robust and versatile digital twin at multicellular or tissue scales, aiming to comprehensively study and predict patient responses to radiation therapy.
Current methods for pathology image segmentation typically treat 2D slices
independently, ignoring valuable cross-slice information. We present
PathSeqSAM, a novel approach that treats 2D pathology slices as sequential
video frames using SAM2's memory mechanisms. Our method introduces a
distance-aware attention mechanism that accounts for variable physical
distances between slices and employs LoRA for domain adaptation. Evaluated on
the KPI Challenge 2024 dataset for glomeruli segmentation, PathSeqSAM
demonstrates improved segmentation quality, particularly in challenging cases
that benefit from cross-slice context. We have publicly released our code at
this https URL
Shanghai Artificial Intelligence LaboratorySouth China Normal UniversityUniversity of GenevaUtrecht UniversityAGH University of KrakowUniversity Medical Center UtrechtMuroran Institute of TechnologyUniversity of Nebraska Medical CenterPrincess Máxima Center for Pediatric OncologyHES-SO Valais-WallisNiigata University of Health and WelfareLa Fe Health Research InstituteWageningen Food Safety ResearchOsaka University Graduate School of MedicineCancer Research Center
Surgery plays an important role within the treatment for neuroblastoma, a
common pediatric cancer. This requires careful planning, often via magnetic
resonance imaging (MRI)-based anatomical 3D models. However, creating these
models is often time-consuming and user dependent. We organized the Surgical
Planning in Pediatric Neuroblastoma (SPPIN) challenge, to stimulate
developments on this topic, and set a benchmark for fully automatic
segmentation of neuroblastoma on multi-model MRI. The challenge started with a
training phase, where teams received 78 sets of MRI scans from 34 patients,
consisting of both diagnostic and post-chemotherapy MRI scans. The final test
phase, consisting of 18 MRI sets from 9 patients, determined the ranking of the
teams. Ranking was based on the Dice similarity coefficient (Dice score), the
95th percentile of the Hausdorff distance (HD95) and the volumetric similarity
(VS). The SPPIN challenge was hosted at MICCAI 2023. The final leaderboard
consisted of 9 teams. The highest-ranking team achieved a median Dice score
0.82, a median HD95 of 7.69 mm and a VS of 0.91, utilizing a large, pretrained
network called STU-Net. A significant difference for the segmentation results
between diagnostic and post-chemotherapy MRI scans was observed (Dice = 0.89 vs
Dice = 0.59, P = 0.01) for the highest-ranking team. SPPIN is the first medical
segmentation challenge in extracranial pediatric oncology. The highest-ranking
team used a large pre-trained network, suggesting that pretraining can be of
use in small, heterogenous datasets. Although the results of the
highest-ranking team were high for most patients, segmentation especially in
small, pre-treated tumors were insufficient. Therefore, more reliable
segmentation methods are needed to create clinically applicable models to aid
surgical planning in pediatric neuroblastoma.
23 Jun 2025
This paper presents a prototype for drug repurposing that integrates quantum-inspired feature extraction with a large language model to analyze multi-omics and clinical data. The system demonstrates improved classification accuracy and more distinct patient clustering compared to classical methods, providing interpretable drug recommendations.
25 Jun 2025
Accurate prediction of cancer type and primary tumor site is critical for effective diagnosis, personalized treatment, and improved outcomes. Traditional models struggle with the complexity of genomic and clinical data, but quantum computing offers enhanced computational capabilities. This study develops a hybrid quantum-classical transformer model, incorporating quantum attention mechanisms via variational quantum circuits (VQCs) to improve prediction accuracy. Using 30,000 anonymized cancer samples from the Genome Warehouse (GWH), data preprocessing included cleaning, encoding, and feature selection. Classical self-attention modules were replaced with quantum attention layers, with classical data encoded into quantum states via amplitude encoding. The model, trained using hybrid backpropagation and quantum gradient calculations, outperformed the classical transformer model, achieving 92.8% accuracy and an AUC of 0.96 compared to 87.5% accuracy and an AUC of 0.89. It also demonstrated 35% faster training and 25% fewer parameters, highlighting computational efficiency. These findings showcase the potential of quantum-enhanced transformers to advance biomedical data analysis, enabling more accurate diagnostics and personalized medicine.
HIV epidemiological data is increasingly complex, requiring advanced computation for accurate cluster detection and forecasting. We employed quantum-accelerated machine learning to analyze HIV prevalence at the ZIP-code level using AIDSVu and synthetic SDoH data for 2022. Our approach compared classical clustering (DBSCAN, HDBSCAN) with a quantum approximate optimization algorithm (QAOA), developed a hybrid quantum-classical neural network for HIV prevalence forecasting, and used quantum Bayesian networks to explore causal links between SDoH factors and HIV incidence. The QAOA-based method achieved 92% accuracy in cluster detection within 1.6 seconds, outperforming classical algorithms. Meanwhile, the hybrid quantum-classical neural network predicted HIV prevalence with 94% accuracy, surpassing a purely classical counterpart. Quantum Bayesian analysis identified housing instability as a key driver of HIV cluster emergence and expansion, with stigma exerting a geographically variable influence. These quantum-enhanced methods deliver greater precision and efficiency in HIV surveillance while illuminating critical causal pathways. This work can guide targeted interventions, optimize resource allocation for PrEP, and address structural inequities fueling HIV transmission.
07 Oct 2025
Statistical significance testing of neural coherence is essential for distinguishing genuine cross-signal coupling from spurious correlations. A widely accepted approach uses surrogate-based inference, where null distributions are generated via time-shift or phase-randomization procedures. While effective, these methods are computationally expensive and yield discrete p-values that can be unstable near decision thresholds, limiting scalability to large EEG/iEEG datasets. We introduce and validate a parametric alternative based on a generalized linear model (GLM) applied to complex-valued time--frequency coefficients (e.g., from DBT or STFT), using a likelihood-ratio test. Using real respiration belt traces as a driver and simulated neural signals contaminated with broadband Gaussian noise, we perform dense sweeps of ground-truth coherence and compare GLM-based inference against time-shift/phase-randomized surrogate testing under matched conditions. GLM achieved comparable or superior sensitivity while producing continuous, stable p-values and a substantial computational advantage. At 80% detection power, GLM detects at C=0.25, whereas surrogate testing requires C=0.49, corresponding to an approximately 6--7 dB SNR improvement. Runtime benchmarking showed GLM to be nearly 200x faster than surrogate approaches. These results establish GLM-based inference on complex time--frequency coefficients as a robust, scalable alternative to surrogate testing, enabling efficient analysis of large EEG/iEEG datasets across channels, frequencies, and participants.
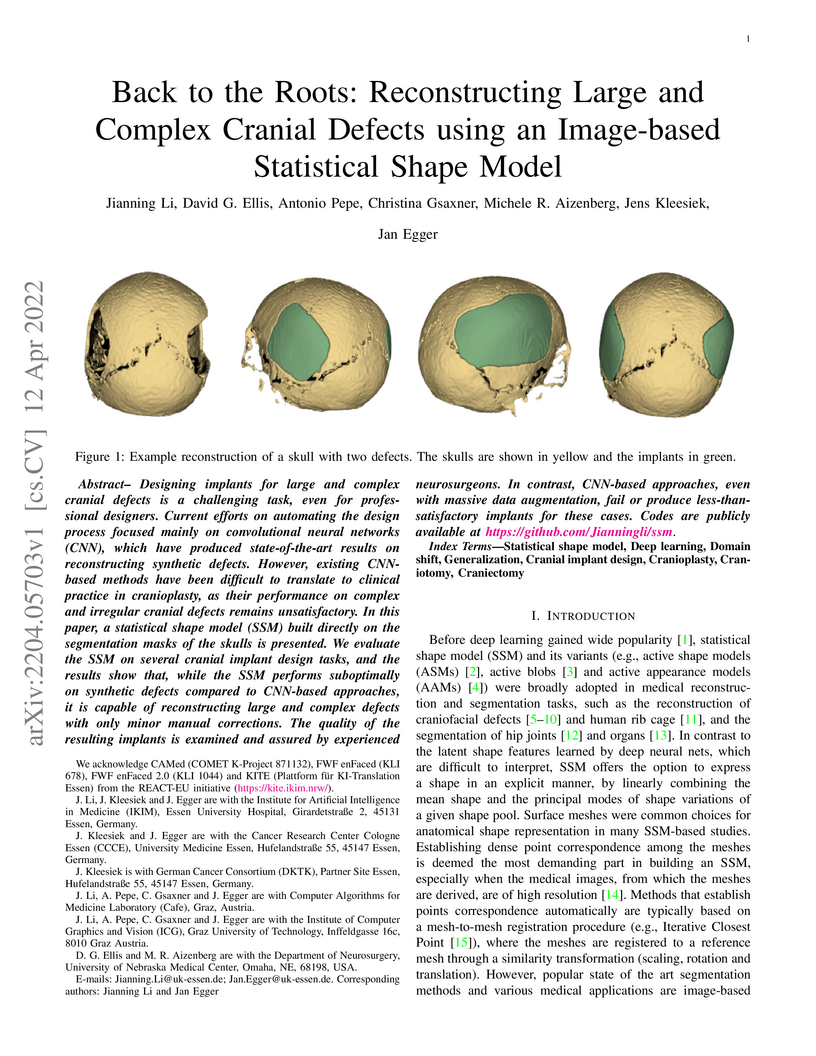
Designing implants for large and complex cranial defects is a challenging task, even for professional designers. Current efforts on automating the design process focused mainly on convolutional neural networks (CNN), which have produced state-of-the-art results on reconstructing synthetic defects. However, existing CNN-based methods have been difficult to translate to clinical practice in cranioplasty, as their performance on complex and irregular cranial defects remains unsatisfactory. In this paper, a statistical shape model (SSM) built directly on the segmentation masks of the skulls is presented. We evaluate the SSM on several cranial implant design tasks, and the results show that, while the SSM performs suboptimally on synthetic defects compared to CNN-based approaches, it is capable of reconstructing large and complex defects with only minor manual corrections. The quality of the resulting implants is examined and assured by experienced neurosurgeons. In contrast, CNN-based approaches, even with massive data augmentation, fail or produce less-than-satisfactory implants for these cases. Codes are publicly available at this https URL
03 Jun 2025
When hypotheses are tested in a stream and real-time decision-making is needed, online sequential hypothesis testing procedures are needed. Furthermore, these hypotheses are commonly partitioned into groups by their nature. For example, the RNA nanocapsules can be partitioned based on therapeutic nucleic acids (siRNAs) being used, as well as the delivery nanocapsules. When selecting effective RNA nanocapsules, simultaneous false discovery rate control at multiple partition levels is needed. In this paper, we develop hypothesis testing procedures which controls false discovery rate (FDR) simultaneously for multiple partitions of hypotheses in an online fashion. We provide rigorous proofs on their FDR or modified FDR (mFDR) control properties and use extensive simulations to demonstrate their performance.
Functional connectivity (FC) has become a primary means of understanding
brain functions by identifying brain network interactions and, ultimately, how
those interactions produce cognitions. A popular definition of FC is by
statistical associations between measured brain regions. However, this could be
problematic since the associations can only provide spatial connections but not
causal interactions among regions of interests. Hence, it is necessary to study
their causal relationship. Directed acyclic graph (DAG) models have been
applied in recent FC studies but often encountered problems such as limited
sample sizes and large number of variables (namely high-dimensional problems),
which lead to both computational difficulty and convergence issues. As a
result, the use of DAG models is problematic, where the identification of DAG
models in general is nondeterministic polynomial time hard (NP-hard). To this
end, we propose a ψ-learning incorporated linear non-Gaussian acyclic
model (ψ-LiNGAM). We use the association model (ψ-learning) to
facilitate causal inferences and the model works well especially for
high-dimensional cases. Our simulation results demonstrate that the proposed
method is more robust and accurate than several existing ones in detecting
graph structure and direction. We then applied it to the resting state fMRI
(rsfMRI) data obtained from the publicly available Philadelphia
Neurodevelopmental Cohort (PNC) to study the cognitive variance, which includes
855 individuals aged 8-22 years. Therein, we have identified three types of hub
structure: the in-hub, out-hub and sum-hub, which correspond to the centers of
receiving, sending and relaying information, respectively. We also detected 16
most important pairs of causal flows. Several of the results have been verified
to be biologically significant.
There are no more papers matching your filters at the moment.