Ask or search anything...
This paper introduces TITAN, a multimodal foundation model that effectively processes whole slide pathology images and text through a three-stage pretraining approach
View blogHEST-1k introduces a large, meticulously curated dataset of paired spatial transcriptomics and histology data, alongside a supporting library and benchmark, enabling the evaluation of deep learning models for predicting gene expression from tissue morphology. This resource helps advance the capabilities of foundation models in pathology and facilitates the exploration of morphomolecular relationships.
View blogResearchers developed VORTEX, an artificial intelligence framework that predicts 3D spatial gene expression patterns across entire tissue volumes using 3D morphological imaging data and limited 2D spatial transcriptomics. This method successfully mapped complex expression landscapes and tumor microenvironments in various cancer types, overcoming the limitations of traditional 2D spatial transcriptomics.
View blogResearchers from Harvard, Mass General Brigham, MIT, and collaborating institutions introduce MedBrowseComp, a benchmark that evaluates AI agents' ability to navigate live medical knowledge bases and synthesize multi-hop evidence from sources like HemOnc.org, PubMed, and ClinicalTrials.gov, revealing that frontier agentic systems achieve as low as 10% accuracy on complex medical information retrieval tasks with performance degrading monotonically as navigation depth increases, while Computer Use Agents show superior performance when starting from domain-specific pages rather than general search engines.
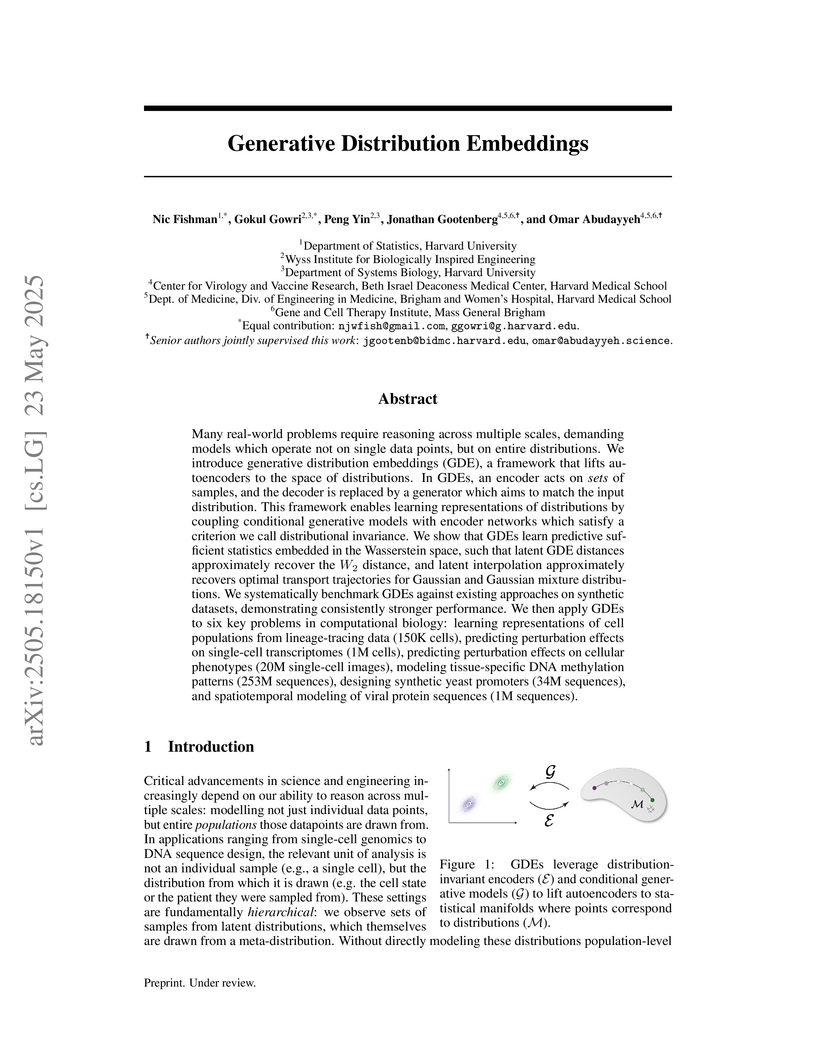
View blogHarvard University and affiliated researchers develop Generative Distribution Embeddings (GDEs), a framework that combines distribution-invariant encoders with conditional generative models to learn representations of probability distributions, demonstrating through systematic benchmarking that GDEs outperform Kernel Mean Embeddings and Wasserstein Wormhole on synthetic datasets while achieving successful applications across six computational biology problems including cell population modeling, perturbation effect prediction, DNA methylation pattern analysis, synthetic promoter design, and viral protein sequence modeling.
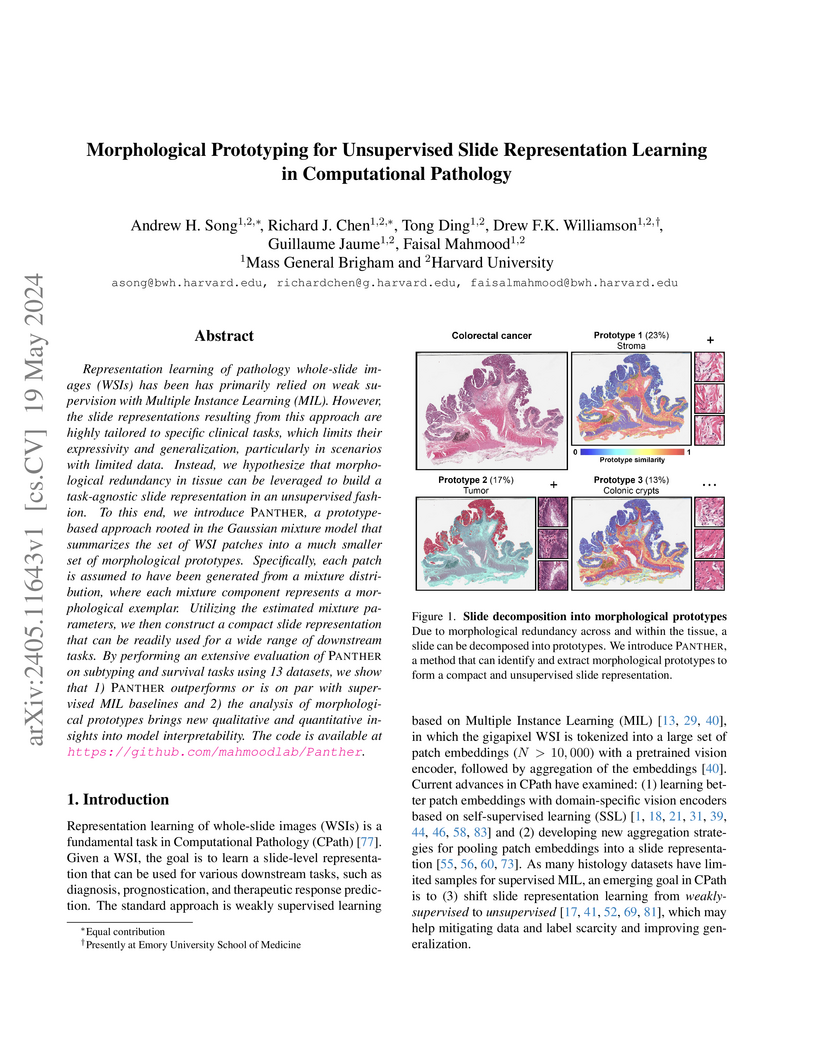
View blogPANTHER introduces an unsupervised framework for learning whole-slide image representations in computational pathology, leveraging Gaussian Mixture Models to capture morphological prototypes and their prevalence. This approach generates task-agnostic slide embeddings that perform comparably to or better than supervised methods across 13 diverse diagnostic and prognostic tasks, offering inherent interpretability.
View blogResearchers discovered that while Large Reasoning Models (LRMs) can be prompted to generate thinking steps in non-English languages, doing so consistently reduces their accuracy on complex math and science problems. They introduced a new challenging multilingual benchmark, XReasoning, to evaluate this trade-off, showing a clear performance degradation despite improved language alignment from prompt modifications or targeted fine-tuning.
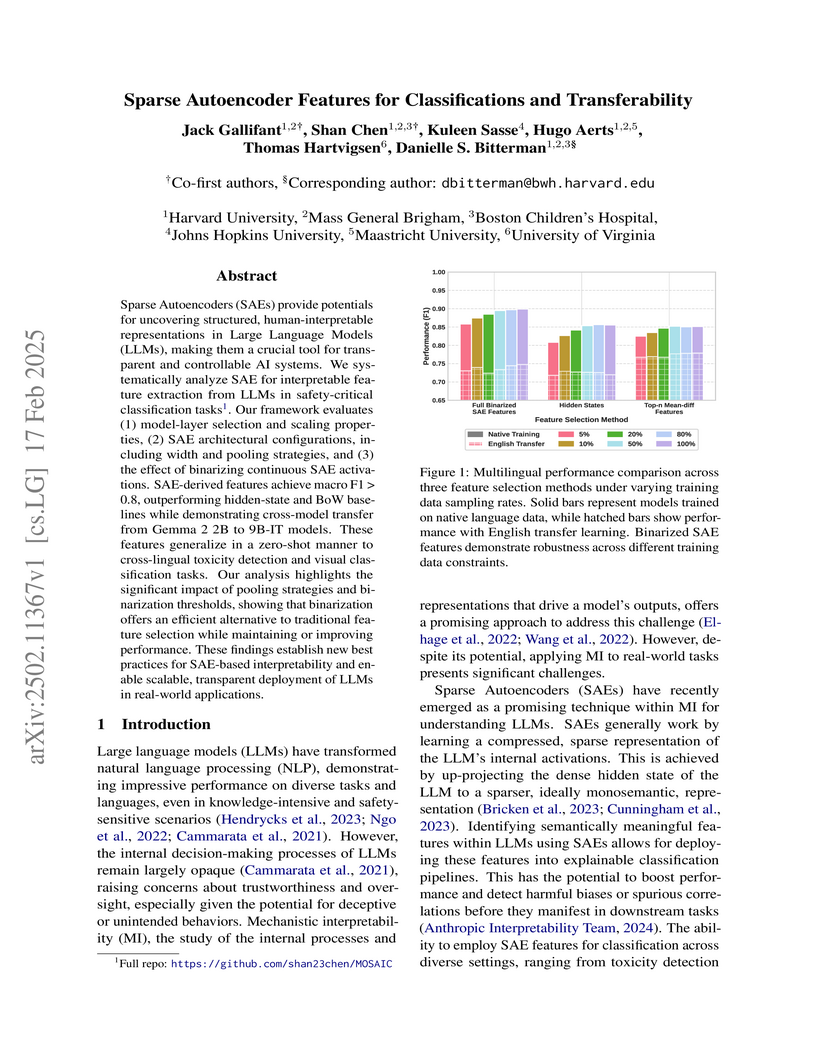
View blogThis research systematically investigates Sparse Autoencoders (SAEs) for extracting interpretable features from Gemma 2 models, establishing benchmarks for SAE-based classification and evaluating their transferability across languages and modalities. It demonstrates that SAE-derived features consistently outperform baselines for various classification tasks.
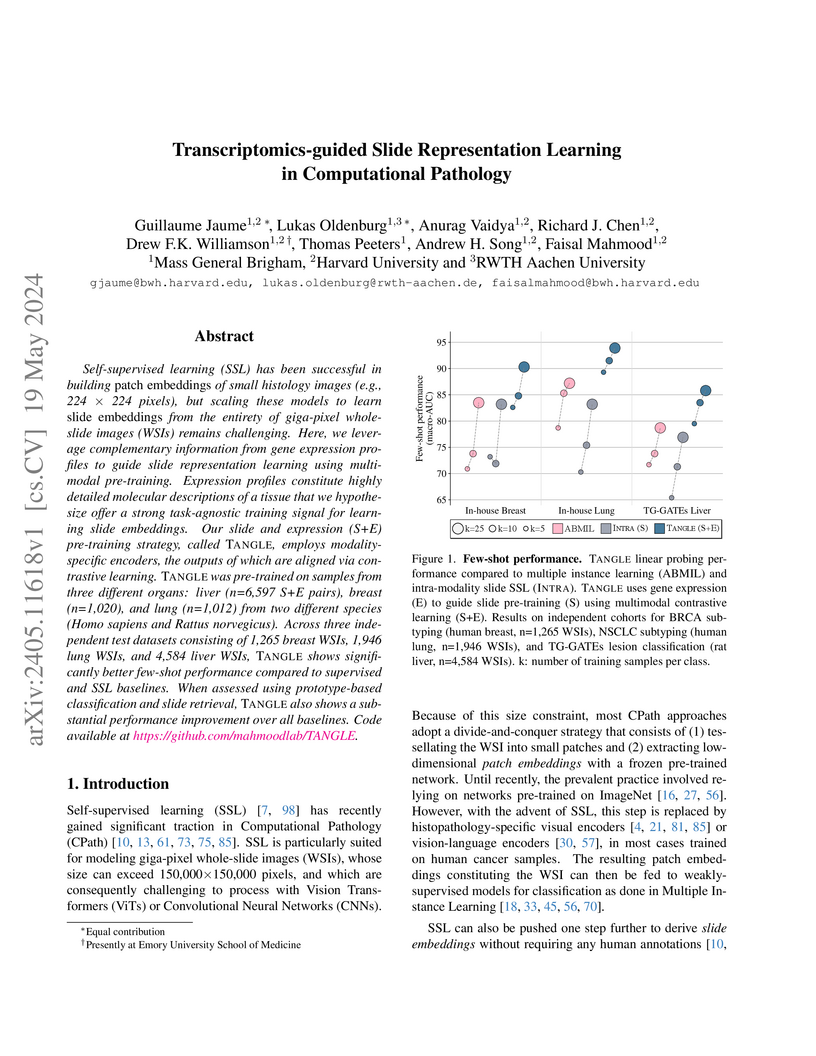
View blogThis research introduces TANGLE, a self-supervised learning framework that guides the creation of high-quality, generalized whole-slide image embeddings using paired transcriptomics data. The framework, applied to both human cancer and rat toxicological pathology, achieved superior performance in few-shot classification, slide retrieval, and demonstrated biologically interpretable representations, including the development of iBOT-Tox, the first large-scale SSL model for rodent histology.
View blog Harvard University
Harvard University the University of Tokyo
the University of Tokyo
 ETH Zurich
ETH Zurich
 Technical University of Munich
Technical University of Munich
 University of Washington
University of Washington University of Cambridge
University of Cambridge
 Johns Hopkins University
Johns Hopkins University

 University of Amsterdam
University of Amsterdam
 Google
Google



 MIT
MIT


 University of Oxford
University of Oxford Microsoft
Microsoft
 Imperial College London
Imperial College London Fudan University
Fudan University