Recursion
Recursion's Valence Labs outlines a vision for 'virtual cells' that can predict, explain, and discover cellular responses to perturbations, leveraging AI/ML on massive interventional datasets. This approach aims to accelerate drug discovery by enabling in silico hypothesis generation and testing without requiring full mechanistic simulations.
TxPert is introduced as a deep learning framework by Valence Labs and Recursion that integrates biochemical knowledge graphs with Graph Neural Networks for out-of-distribution transcriptomic perturbation prediction. The system demonstrates state-of-the-art performance, approaching experimental reproducibility for single perturbations in known cell types and generalizing to novel double perturbations and unseen cell lines.
Researchers at Mila and collaborators introduce Relative Trajectory Balance (RTB), a novel objective for training diffusion models to perform amortized sampling of intractable posterior distributions. This method offers an asymptotically unbiased approach for posterior inference, demonstrating its effectiveness across tasks in vision, language, and continuous control.
Understanding the relationships among genes, compounds, and their
interactions in living organisms remains limited due to technological
constraints and the complexity of biological data. Deep learning has shown
promise in exploring these relationships using various data types. However,
transcriptomics, which provides detailed insights into cellular states, is
still underused due to its high noise levels and limited data availability.
Recent advancements in transcriptomics sequencing provide new opportunities to
uncover valuable insights, especially with the rise of many new foundation
models for transcriptomics, yet no benchmark has been made to robustly evaluate
the effectiveness of these rising models for perturbation analysis. This
article presents a novel biologically motivated evaluation framework and a
hierarchy of perturbation analysis tasks for comparing the performance of
pretrained foundation models to each other and to more classical techniques of
learning from transcriptomics data. We compile diverse public datasets from
different sequencing techniques and cell lines to assess models performance.
Our approach identifies scVI and PCA to be far better suited models for
understanding biological perturbations in comparison to existing foundation
models, especially in their application in real-world scenarios.
We study the problem of generating diverse candidates in the context of Multi-Objective Optimization. In many applications of machine learning such as drug discovery and material design, the goal is to generate candidates which simultaneously optimize a set of potentially conflicting objectives. Moreover, these objectives are often imperfect evaluations of some underlying property of interest, making it important to generate diverse candidates to have multiple options for expensive downstream evaluations. We propose Multi-Objective GFlowNets (MOGFNs), a novel method for generating diverse Pareto optimal solutions, based on GFlowNets. We introduce two variants of MOGFNs: MOGFN-PC, which models a family of independent sub-problems defined by a scalarization function, with reward-conditional GFlowNets, and MOGFN-AL, which solves a sequence of sub-problems defined by an acquisition function in an active learning loop. Our experiments on wide variety of synthetic and benchmark tasks demonstrate advantages of the proposed methods in terms of the Pareto performance and importantly, improved candidate diversity, which is the main contribution of this work.
Local Search GFlowNets (LS-GFN) integrates a local search mechanism into Generative Flow Networks, enhancing their capacity to consistently generate diverse and high-rewarding samples. This approach demonstrates improved reward distribution matching, increased mode discovery, and superior diversity compared to current GFlowNet methods and reward-maximization baselines across various biochemical tasks.
Amortized inference is the task of training a parametric model, such as a
neural network, to approximate a distribution with a given unnormalized density
where exact sampling is intractable. When sampling is implemented as a
sequential decision-making process, reinforcement learning (RL) methods, such
as generative flow networks, can be used to train the sampling policy.
Off-policy RL training facilitates the discovery of diverse, high-reward
candidates, but existing methods still face challenges in efficient
exploration. We propose to use an adaptive training distribution (the \teacher)
to guide the training of the primary amortized sampler (the \student). The
\teacher, an auxiliary behavior model, is trained to sample high-loss regions
of the \student and can generalize across unexplored modes, thereby enhancing
mode coverage by providing an efficient training curriculum. We validate the
effectiveness of this approach in a synthetic environment designed to present
an exploration challenge, two diffusion-based sampling tasks, and four
biochemical discovery tasks demonstrating its ability to improve sample
efficiency and mode coverage. Source code is available at
this https URL
Researchers from Mila, KAIST, and Valence Labs developed δ-Conservative Search (δ-CS), an off-policy reinforcement learning method that enhances Generative Flow Networks (GFlowNets) for biological sequence design. This approach mitigates proxy model unreliability on out-of-distribution inputs, achieving superior performance on diverse tasks including DNA, RNA, and protein design with faster convergence and higher final scores compared to existing baselines.
This work introduces a framework for solving Bayesian inverse problems by leveraging Relative Trajectory Balance (RTB) with off-policy reinforcement learning to fine-tune diffusion models. The method enables more accurate and unbiased sampling from complex posterior distributions, demonstrated on image reconstruction tasks and scientific applications such as gravitational lensing.
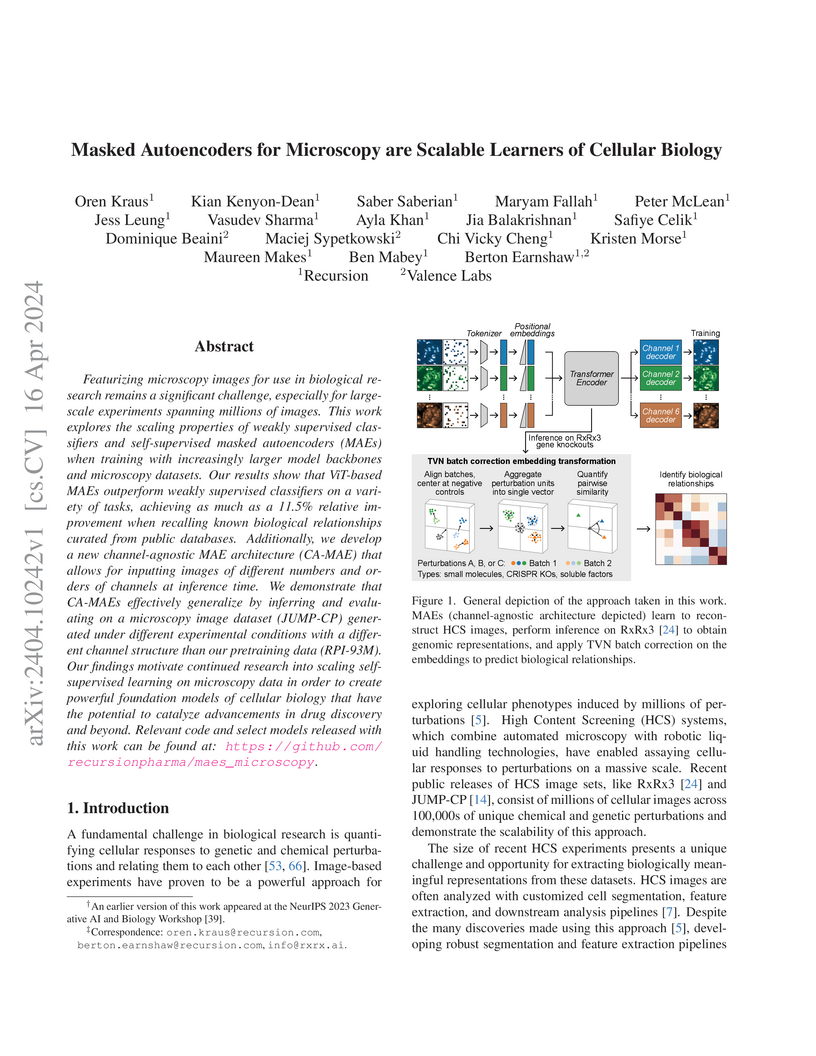
Featurizing microscopy images for use in biological research remains a
significant challenge, especially for large-scale experiments spanning millions
of images. This work explores the scaling properties of weakly supervised
classifiers and self-supervised masked autoencoders (MAEs) when training with
increasingly larger model backbones and microscopy datasets. Our results show
that ViT-based MAEs outperform weakly supervised classifiers on a variety of
tasks, achieving as much as a 11.5% relative improvement when recalling known
biological relationships curated from public databases. Additionally, we
develop a new channel-agnostic MAE architecture (CA-MAE) that allows for
inputting images of different numbers and orders of channels at inference time.
We demonstrate that CA-MAEs effectively generalize by inferring and evaluating
on a microscopy image dataset (JUMP-CP) generated under different experimental
conditions with a different channel structure than our pretraining data
(RPI-93M). Our findings motivate continued research into scaling
self-supervised learning on microscopy data in order to create powerful
foundation models of cellular biology that have the potential to catalyze
advancements in drug discovery and beyond.
Models that accurately predict properties based on chemical structure are valuable tools in drug discovery. However, for many properties, public and private training sets are typically small, and it is difficult for the models to generalize well outside of the training data. Recently, large language models have addressed this problem by using self-supervised pretraining on large unlabeled datasets, followed by fine-tuning on smaller, labeled datasets. In this paper, we report MolE, a molecular foundation model that adapts the DeBERTa architecture to be used on molecular graphs together with a two-step pretraining strategy. The first step of pretraining is a self-supervised approach focused on learning chemical structures, and the second step is a massive multi-task approach to learn biological information. We show that fine-tuning pretrained MolE achieves state-of-the-art results on 9 of the 22 ADMET tasks included in the Therapeutic Data Commons.
Generating stable molecular conformations is crucial in several drug discovery applications, such as estimating the binding affinity of a molecule to a target. Recently, generative machine learning methods have emerged as a promising, more efficient method than molecular dynamics for sampling of conformations from the Boltzmann distribution. In this paper, we introduce Torsional-GFN, a conditional GFlowNet specifically designed to sample conformations of molecules proportionally to their Boltzmann distribution, using only a reward function as training signal. Conditioned on a molecular graph and its local structure (bond lengths and angles), Torsional-GFN samples rotations of its torsion angles. Our results demonstrate that Torsional-GFN is able to sample conformations approximately proportional to the Boltzmann distribution for multiple molecules with a single model, and allows for zero-shot generalization to unseen bond lengths and angles coming from the MD simulations for such molecules. Our work presents a promising avenue for scaling the proposed approach to larger molecular systems, achieving zero-shot generalization to unseen molecules, and including the generation of the local structure into the GFlowNet model.
A novel sparse dictionary learning algorithm, Iterative Codebook Feature Learning (ICFL) combined with PCA whitening, extracts biologically meaningful and interpretable concepts from unsupervised microscopy vision foundation models. This approach achieves feature selectivity comparable to handcrafted features and enables single-cell level interpretability, revealing channel-specific morphological changes and differentiating cell subpopulations.
Pairwise interactions between perturbations to a system can provide evidence
for the causal dependencies of the underlying underlying mechanisms of a
system. When observations are low dimensional, hand crafted measurements,
detecting interactions amounts to simple statistical tests, but it is not
obvious how to detect interactions between perturbations affecting latent
variables. We derive two interaction tests that are based on pairwise
interventions, and show how these tests can be integrated into an active
learning pipeline to efficiently discover pairwise interactions between
perturbations. We illustrate the value of these tests in the context of
biology, where pairwise perturbation experiments are frequently used to reveal
interactions that are not observable from any single perturbation. Our tests
can be run on unstructured data, such as the pixels in an image, which enables
a more general notion of interaction than typical cell viability experiments,
and can be run on cheaper experimental assays. We validate on several synthetic
and real biological experiments that our tests are able to identify interacting
pairs effectively. We evaluate our approach on a real biological experiment
where we knocked out 50 pairs of genes and measured the effect with microscopy
images. We show that we are able to recover significantly more known biological
interactions than random search and standard active learning baselines.
Researchers at Mila - Quebec AI Institute developed ACTIONPIECE, a method that automatically identifies and incorporates frequently occurring action sequences into an agent's action space. This approach improves performance in sequential decision-making tasks, leading to faster discovery of high-reward solutions and better density estimation in generative models, particularly GFlowNets.
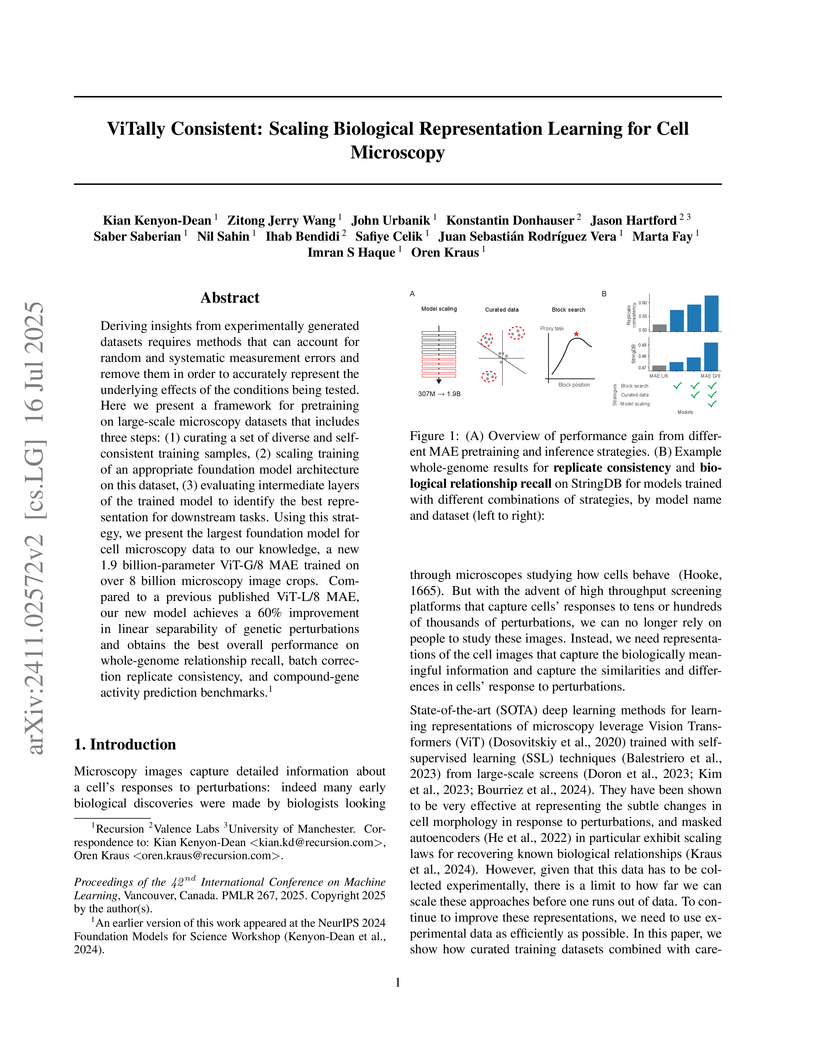
Researchers at Recursion developed a framework for biological representation learning in cell microscopy, combining a curated dataset (Phenoprints-16M) with a 1.9 billion-parameter Vision Transformer and a method to select optimal intermediate layers. This approach achieved state-of-the-art performance, including a 60% improvement in genetic perturbation separability and a 42% relative improvement in compound-gene activity prediction.
High-throughput screening techniques are commonly used to obtain large quantities of data in many fields of biology. It is well known that artifacts arising from variability in the technical execution of different experimental batches within such screens confound these observations and can lead to invalid biological conclusions. It is therefore necessary to account for these batch effects when analyzing outcomes. In this paper we describe RxRx1, a biological dataset designed specifically for the systematic study of batch effect correction methods. The dataset consists of 125,510 high-resolution fluorescence microscopy images of human cells under 1,138 genetic perturbations in 51 experimental batches across 4 cell types. Visual inspection of the images alone clearly demonstrates significant batch effects. We propose a classification task designed to evaluate the effectiveness of experimental batch correction methods on these images and examine the performance of a number of correction methods on this task. Our goal in releasing RxRx1 is to encourage the development of effective experimental batch correction methods that generalize well to unseen experimental batches. The dataset can be downloaded at this https URL.
High Content Screening (HCS) microscopy datasets have transformed the ability
to profile cellular responses to genetic and chemical perturbations, enabling
cell-based inference of drug-target interactions (DTI). However, the adoption
of representation learning methods for HCS data has been hindered by the lack
of accessible datasets and robust benchmarks. To address this gap, we present
RxRx3-core, a curated and compressed subset of the RxRx3 dataset, and an
associated DTI benchmarking task. At just 18GB, RxRx3-core significantly
reduces the size barrier associated with large-scale HCS datasets while
preserving critical data necessary for benchmarking representation learning
models against a zero-shot DTI prediction task. RxRx3-core includes 222,601
microscopy images spanning 736 CRISPR knockouts and 1,674 compounds at 8
concentrations. RxRx3-core is available on HuggingFace and Polaris, along with
pre-trained embeddings and benchmarking code, ensuring accessibility for the
research community. By providing a compact dataset and robust benchmarks, we
aim to accelerate innovation in representation learning methods for HCS data
and support the discovery of novel biological insights.
Understanding cellular responses to stimuli is crucial for biological
discovery and drug development. Transcriptomics provides interpretable,
gene-level insights, while microscopy imaging offers rich predictive features
but is harder to interpret. Weakly paired datasets, where samples share
biological states, enable multimodal learning but are scarce, limiting their
utility for training and multimodal inference. We propose a framework to
enhance transcriptomics by distilling knowledge from microscopy images. Using
weakly paired data, our method aligns and binds modalities, enriching gene
expression representations with morphological information. To address data
scarcity, we introduce (1) Semi-Clipped, an adaptation of CLIP for cross-modal
distillation using pretrained foundation models, achieving state-of-the-art
results, and (2) PEA (Perturbation Embedding Augmentation), a novel
augmentation technique that enhances transcriptomics data while preserving
inherent biological information. These strategies improve the predictive power
and retain the interpretability of transcriptomics, enabling rich unimodal
representations for complex biological tasks.
Generative chemical language models (CLMs) have demonstrated strong capabilities in molecular design, yet their impact in drug discovery remains limited by the absence of reliable reward signals and the lack of interpretability in their outputs. We present SAFE-T, a generalist chemical modeling framework that conditions on biological context -- such as protein targets or mechanisms of action -- to prioritize and design molecules without relying on structural information or engineered scoring functions. SAFE-T models the conditional likelihood of fragment-based molecular sequences given a biological prompt, enabling principled scoring of molecules across tasks such as virtual screening, drug-target interaction prediction, and activity cliff detection. Moreover, it supports goal-directed generation by sampling from this learned distribution, aligning molecular design with biological objectives. In comprehensive zero-shot evaluations across predictive (LIT-PCBA, DAVIS, KIBA, ACNet) and generative (DRUG, PMO) benchmarks, SAFE-T consistently achieves performance comparable to or better than existing approaches while being significantly faster. Fragment-level attribution further reveals that SAFE-T captures known structure-activity relationships, supporting interpretable and biologically grounded design. Together with its computational efficiency, these results demonstrate that conditional generative CLMs can unify scoring and generation to accelerate early-stage drug discovery.
There are no more papers matching your filters at the moment.