University Hospital Erlangen
How does the mind organize thoughts? The hippocampal-entorhinal complex is
thought to support domain-general representation and processing of structural
knowledge of arbitrary state, feature and concept spaces. In particular, it
enables the formation of cognitive maps, and navigation on these maps, thereby
broadly contributing to cognition. It has been proposed that the concept of
multi-scale successor representations provides an explanation of the underlying
computations performed by place and grid cells. Here, we present a neural
network based approach to learn such representations, and its application to
different scenarios: a spatial exploration task based on supervised learning, a
spatial navigation task based on reinforcement learning, and a non-spatial task
where linguistic constructions have to be inferred by observing sample
sentences. In all scenarios, the neural network correctly learns and
approximates the underlying structure by building successor representations.
Furthermore, the resulting neural firing patterns are strikingly similar to
experimentally observed place and grid cell firing patterns. We conclude that
cognitive maps and neural network-based successor representations of structured
knowledge provide a promising way to overcome some of the short comings of deep
learning towards artificial general intelligence.
Refusal behavior in large language models (LLMs) enables them to decline responding to harmful, unethical, or inappropriate prompts, ensuring alignment with ethical standards. This paper investigates refusal behavior across six LLMs from three architectural families. We challenge the assumption of refusal as a linear phenomenon by employing dimensionality reduction techniques, including PCA, t-SNE, and UMAP. Our results reveal that refusal mechanisms exhibit nonlinear, multidimensional characteristics that vary by model architecture and layer. These findings highlight the need for nonlinear interpretability to improve alignment research and inform safer AI deployment strategies.
13 Oct 2025
Clinical decision-making in radiology increasingly benefits from artificial intelligence (AI), particularly through large language models (LLMs). However, traditional retrieval-augmented generation (RAG) systems for radiology question answering (QA) typically rely on single-step retrieval, limiting their ability to handle complex clinical reasoning tasks. Here we propose radiology Retrieval and Reasoning (RaR), a multi-step retrieval and reasoning framework designed to improve diagnostic accuracy, factual consistency, and clinical reliability of LLMs in radiology question answering. We evaluated 25 LLMs spanning diverse architectures, parameter scales (0.5B to >670B), and training paradigms (general-purpose, reasoning-optimized, clinically fine-tuned), using 104 expert-curated radiology questions from previously established RSNA-RadioQA and ExtendedQA datasets. To assess generalizability, we additionally tested on an unseen internal dataset of 65 real-world radiology board examination questions. RaR significantly improved mean diagnostic accuracy over zero-shot prompting and conventional online RAG. The greatest gains occurred in small-scale models, while very large models (>200B parameters) demonstrated minimal changes (<2% improvement). Additionally, RaR retrieval reduced hallucinations (mean 9.4%) and retrieved clinically relevant context in 46% of cases, substantially aiding factual grounding. Even clinically fine-tuned models showed gains from RaR (e.g., MedGemma-27B), indicating that retrieval remains beneficial despite embedded domain knowledge. These results highlight the potential of RaR to enhance factuality and diagnostic accuracy in radiology QA, warranting future studies to validate their clinical utility. All datasets, code, and the full RaR framework are publicly available to support open research and clinical translation.
10 Jun 2025
Predictive coding theory suggests that the brain continuously anticipates
upcoming words to optimize language processing, but the neural mechanisms
remain unclear, particularly in naturalistic speech. Here, we simultaneously
recorded EEG and MEG data from 29 participants while they listened to an audio
book and assigned predictability scores to nouns using the BERT language model.
Our results show that higher predictability is associated with reduced neural
responses during word recognition, as reflected in lower N400 amplitudes, and
with increased anticipatory activity before word onset. EEG data revealed
increased pre-activation in left fronto-temporal regions, while MEG showed a
tendency for greater sensorimotor engagement in response to low-predictability
words, suggesting a possible motor-related component to linguistic
anticipation. These findings provide new evidence that the brain dynamically
integrates top-down predictions with bottom-up sensory input to facilitate
language comprehension. To our knowledge, this is the first study to
demonstrate these effects using naturalistic speech stimuli, bridging
computational language models with neurophysiological data. Our findings
provide novel insights for cognitive computational neuroscience, advancing the
understanding of predictive processing in language and inspiring the
development of neuroscience-inspired AI. Future research should explore the
role of prediction and sensory precision in shaping neural responses and
further refine models of language processing.
Survival prediction for cancer patients is critical for optimal treatment
selection and patient management. Current patient survival prediction methods
typically extract survival information from patients' clinical record data or
biological and imaging data. In practice, experienced clinicians can have a
preliminary assessment of patients' health status based on patients' observable
physical appearances, which are mainly facial features. However, such
assessment is highly subjective. In this work, the efficacy of objectively
capturing and using prognostic information contained in conventional portrait
photographs using deep learning for survival predication purposes is
investigated for the first time. A pre-trained StyleGAN2 model is fine-tuned on
a custom dataset of our cancer patients' photos to empower its generator with
generative ability suitable for patients' photos. The StyleGAN2 is then used to
embed the photographs to its highly expressive latent space. Utilizing the
state-of-the-art survival analysis models and based on StyleGAN's latent space
photo embeddings, this approach achieved a C-index of 0.677, which is notably
higher than chance and evidencing the prognostic value embedded in simple 2D
facial images. In addition, thanks to StyleGAN's interpretable latent space,
our survival prediction model can be validated for relying on essential facial
features, eliminating any biases from extraneous information like clothing or
background. Moreover, a health attribute is obtained from regression
coefficients, which has important potential value for patient care.
Computed Tomography (CT) image reconstruction is crucial for accurate diagnosis and deep learning approaches have demonstrated significant potential in improving reconstruction quality. However, the choice of loss function profoundly affects the reconstructed images. Traditional mean squared error loss often produces blurry images lacking fine details, while alternatives designed to improve may introduce structural artifacts or other undesirable effects. To address these limitations, we propose Eagle-Loss, a novel loss function designed to enhance the visual quality of CT image reconstructions. Eagle-Loss applies spectral analysis of localized features within gradient changes to enhance sharpness and well-defined edges. We evaluated Eagle-Loss on two public datasets across low-dose CT reconstruction and CT field-of-view extension tasks. Our results show that Eagle-Loss consistently improves the visual quality of reconstructed images, surpassing state-of-the-art methods across various network architectures. Code and data are available at \url{this https URL}.
This study investigates the internal representations of verb-particle
combinations, called multi-word verbs, within transformer-based large language
models (LLMs), specifically examining how these models capture lexical and
syntactic properties at different neural network layers. Using the BERT
architecture, we analyze the representations of its layers for two different
verb-particle constructions: phrasal verbs like 'give up' and prepositional
verbs like 'look at'. Our methodology includes training probing classifiers on
the internal representations to classify these categories at both word and
sentence levels. The results indicate that the model's middle layers achieve
the highest classification accuracies. To further analyze the nature of these
distinctions, we conduct a data separability test using the Generalized
Discrimination Value (GDV). While GDV results show weak linear separability
between the two verb types, probing classifiers still achieve high accuracy,
suggesting that representations of these linguistic categories may be
non-linearly separable. This aligns with previous research indicating that
linguistic distinctions in neural networks are not always encoded in a linearly
separable manner. These findings computationally support usage-based claims on
the representation of verb-particle constructions and highlight the complex
interaction between neural network architectures and linguistic structures.
Understanding the relationship between vocal tract motion during speech and
the resulting acoustic signal is crucial for aided clinical assessment and
developing personalized treatment and rehabilitation strategies. Toward this
goal, we introduce an audio-to-video generation framework for creating Real
Time/cine-Magnetic Resonance Imaging (RT-/cine-MRI) visuals of the vocal tract
from speech signals. Our framework first preprocesses RT-/cine-MRI sequences
and speech samples to achieve temporal alignment, ensuring synchronization
between visual and audio data. We then employ a modified stable diffusion
model, integrating structural and temporal blocks, to effectively capture
movement characteristics and temporal dynamics in the synchronized data. This
process enables the generation of MRI sequences from new speech inputs,
improving the conversion of audio into visual data. We evaluated our framework
on healthy controls and tongue cancer patients by analyzing and comparing the
vocal tract movements in synthesized videos. Our framework demonstrated
adaptability to new speech inputs and effective generalization. In addition,
positive human evaluations confirmed its effectiveness, with realistic and
accurate visualizations, suggesting its potential for outpatient therapy and
personalized simulation of vocal tract visualizations.
Biophysical model fitting plays a key role in obtaining quantitative
parameters from physiological signals and images. However, the model complexity
for molecular magnetic resonance imaging (MRI) often translates into excessive
computation time, which makes clinical use impractical. Here, we present a
generic computational approach for solving the parameter extraction inverse
problem posed by ordinary differential equation (ODE) modeling coupled with
experimental measurement of the system dynamics. This is achieved by
formulating a numerical ODE solver to function as a step-wise analytical one,
thereby making it compatible with automatic differentiation-based optimization.
This enables efficient gradient-based model fitting, and provides a new
approach to parameter quantification based on self-supervised learning from a
single data observation. The neural-network-based train-by-fit pipeline was
used to quantify semisolid magnetization transfer (MT) and chemical exchange
saturation transfer (CEST) amide proton exchange parameters in the human brain,
in an in-vivo molecular MRI study (n = 4). The entire pipeline of the first
whole brain quantification was completed in 18.3 ± 8.3 minutes. Reusing the
single-subject-trained network for inference in new subjects took 1.0 ± 0.2
s, to provide results in agreement with literature values and scan-specific fit
results.
Providing more precise tissue attenuation information, synthetic computed
tomography (sCT) generated from magnetic resonance imaging (MRI) contributes to
improved radiation therapy treatment planning. In our study, we employ the
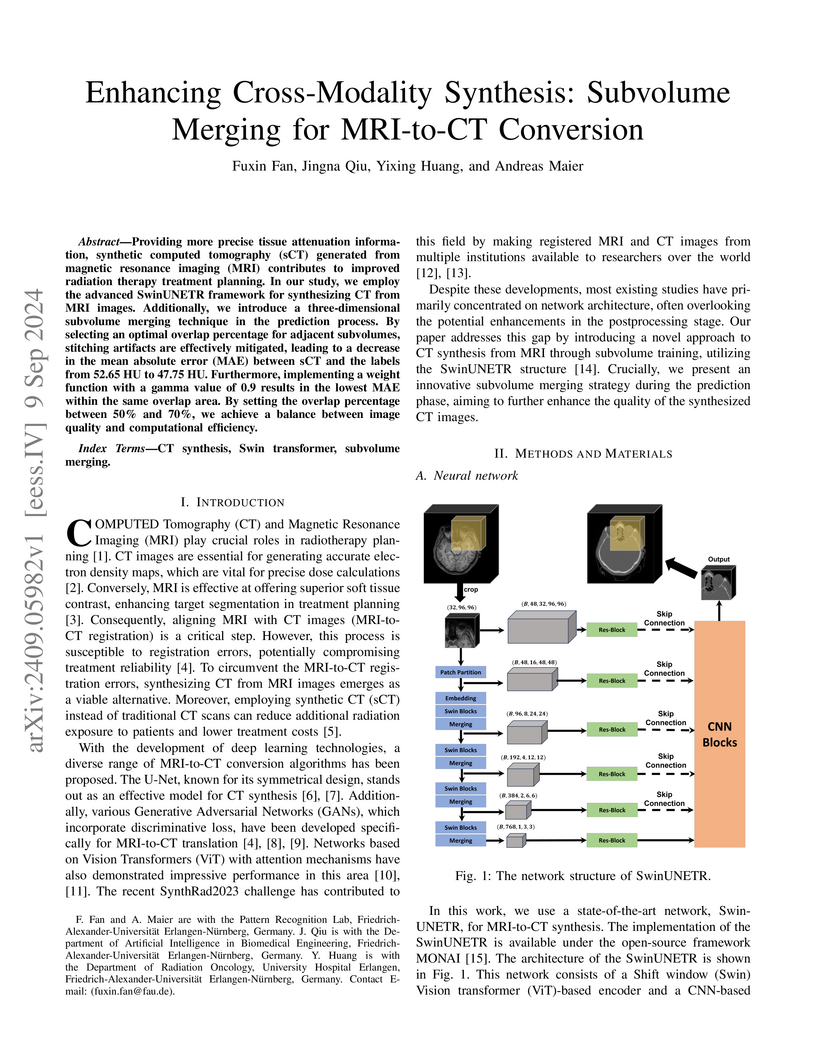
advanced SwinUNETR framework for synthesizing CT from MRI images. Additionally,
we introduce a three-dimensional subvolume merging technique in the prediction
process. By selecting an optimal overlap percentage for adjacent subvolumes,
stitching artifacts are effectively mitigated, leading to a decrease in the
mean absolute error (MAE) between sCT and the labels from 52.65 HU to 47.75 HU.
Furthermore, implementing a weight function with a gamma value of 0.9 results
in the lowest MAE within the same overlap area. By setting the overlap
percentage between 50% and 70%, we achieve a balance between image quality and
computational efficiency.
Researchers at the University Erlangen-Nuremberg developed a neural network model utilizing multi-scale successor representations to construct cognitive maps of abstract semantic spaces, demonstrating how the discount factor regulates map granularity and facilitates the emergence of conceptual hierarchies. This approach also enables robust inference of missing features from partial input, mimicking the brain's capacity to derive context from incomplete information.
Computed tomography is widely used as an imaging tool to visualize three-dimensional structures with expressive bone-soft tissue contrast. However, CT resolution and radiation dose are tightly entangled, highlighting the importance of low-dose CT combined with sophisticated denoising algorithms. Most data-driven denoising techniques are based on deep neural networks and, therefore, contain hundreds of thousands of trainable parameters, making them incomprehensible and prone to prediction failures. Developing understandable and robust denoising algorithms achieving state-of-the-art performance helps to minimize radiation dose while maintaining data integrity. This work presents an open-source CT denoising framework based on the idea of bilateral filtering. We propose a bilateral filter that can be incorporated into a deep learning pipeline and optimized in a purely data-driven way by calculating the gradient flow toward its hyperparameters and its input. Denoising in pure image-to-image pipelines and across different domains such as raw detector data and reconstructed volume, using a differentiable backprojection layer, is demonstrated. Although only using three spatial parameters and one range parameter per filter layer, the proposed denoising pipelines can compete with deep state-of-the-art denoising architectures with several hundred thousand parameters. Competitive denoising performance is achieved on x-ray microscope bone data (0.7053 and 33.10) and the 2016 Low Dose CT Grand Challenge dataset (0.9674 and 43.07) in terms of SSIM and PSNR. Due to the extremely low number of trainable parameters with well-defined effect, prediction reliance and data integrity is guaranteed at any time in the proposed pipelines, in contrast to most other deep learning-based denoising architectures.
The ability to transmit and receive complex information via language is unique to humans and is the basis of traditions, culture and versatile social interactions. Through the disruptive introduction of transformer based large language models (LLMs) humans are not the only entity to "understand" and produce language any more. In the present study, we have performed the first steps to use LLMs as a model to understand fundamental mechanisms of language processing in neural networks, in order to make predictions and generate hypotheses on how the human brain does language processing. Thus, we have used ChatGPT to generate seven different stylistic variations of ten different narratives (Aesop's fables). We used these stories as input for the open source LLM BERT and have analyzed the activation patterns of the hidden units of BERT using multi-dimensional scaling and cluster analysis. We found that the activation vectors of the hidden units cluster according to stylistic variations in earlier layers of BERT (1) than narrative content (4-5). Despite the fact that BERT consists of 12 identical building blocks that are stacked and trained on large text corpora, the different layers perform different tasks. This is a very useful model of the human brain, where self-similar structures, i.e. different areas of the cerebral cortex, can have different functions and are therefore well suited to processing language in a very efficient way. The proposed approach has the potential to open the black box of LLMs on the one hand, and might be a further step to unravel the neural processes underlying human language processing and cognition in general.
Denoising Diffusion Probabilistic models have become increasingly popular due to their ability to offer probabilistic modeling and generate diverse outputs. This versatility inspired their adaptation for image segmentation, where multiple predictions of the model can produce segmentation results that not only achieve high quality but also capture the uncertainty inherent in the model. Here, powerful architectures were proposed for improving diffusion segmentation performance. However, there is a notable lack of analysis and discussions on the differences between diffusion segmentation and image generation, and thorough evaluations are missing that distinguish the improvements these architectures provide for segmentation in general from their benefit for diffusion segmentation specifically. In this work, we critically analyse and discuss how diffusion segmentation for medical images differs from diffusion image generation, with a particular focus on the training behavior. Furthermore, we conduct an assessment how proposed diffusion segmentation architectures perform when trained directly for segmentation. Lastly, we explore how different medical segmentation tasks influence the diffusion segmentation behavior and the diffusion process could be adapted accordingly. With these analyses, we aim to provide in-depth insights into the behavior of diffusion segmentation that allow for a better design and evaluation of diffusion segmentation methods in the future.
In the evolving landscape of data science, the accurate quantification of
clustering in high-dimensional data sets remains a significant challenge,
especially in the absence of predefined labels. This paper introduces a novel
approach, the Entropy of Distance Distribution (EDD), which represents a
paradigm shift in label-free clustering analysis. Traditional methods, reliant
on discrete labels, often struggle to discern intricate cluster patterns in
unlabeled data. EDD, however, leverages the characteristic differences in
pairwise point-to-point distances to discern clustering tendencies, independent
of data labeling.
Our method employs the Shannon information entropy to quantify the
'peakedness' or 'flatness' of distance distributions in a data set. This
entropy measure, normalized against its maximum value, effectively
distinguishes between strongly clustered data (indicated by pronounced peaks in
distance distribution) and more homogeneous, non-clustered data sets. This
label-free quantification is resilient against global translations and
permutations of data points, and with an additional dimension-wise z-scoring,
it becomes invariant to data set scaling.
We demonstrate the efficacy of EDD through a series of experiments involving
two-dimensional data spaces with Gaussian cluster centers. Our findings reveal
a monotonic increase in the EDD value with the widening of cluster widths,
moving from well-separated to overlapping clusters. This behavior underscores
the method's sensitivity and accuracy in detecting varying degrees of
clustering. EDD's potential extends beyond conventional clustering analysis,
offering a robust, scalable tool for unraveling complex data structures without
reliance on pre-assigned labels.
This research analyzes BERT's internal mechanisms, demonstrating its robust capacity to cluster narrative content over subtle individual authorial styles across its layers. The study reveals that BERT's internal representations progressively refine content-based clusters while showing minimal encoding of nuanced stylistic variations introduced via neural style transfer.
Measurement of stride-related, biomechanical parameters is the common rationale for objective gait impairment scoring. State-of-the-art double integration approaches to extract these parameters from inertial sensor data are, however, limited in their clinical applicability due to the underlying assumptions. To overcome this, we present a method to translate the abstract information provided by wearable sensors to context-related expert features based on deep convolutional neural networks. Regarding mobile gait analysis, this enables integration-free and data-driven extraction of a set of 8 spatio-temporal stride parameters. To this end, two modelling approaches are compared: A combined network estimating all parameters of interest and an ensemble approach that spawns less complex networks for each parameter individually. The ensemble approach is outperforming the combined modelling in the current application. On a clinically relevant and publicly available benchmark dataset, we estimate stride length, width and medio-lateral change in foot angle up to −0.15±6.09 cm, −0.09±4.22 cm and 0.13±3.78∘ respectively. Stride, swing and stance time as well as heel and toe contact times are estimated up to ±0.07, ±0.05, ±0.07, ±0.07 and ±0.12 s respectively. This is comparable to and in parts outperforming or defining state-of-the-art. Our results further indicate that the proposed change in methodology could substitute assumption-driven double-integration methods and enable mobile assessment of spatio-temporal stride parameters in clinically critical situations as e.g. in the case of spastic gait impairments.
Deep learning-based image generation has seen significant advancements with diffusion models, notably improving the quality of generated images. Despite these developments, generating images with unseen characteristics beneficial for downstream tasks has received limited attention. To bridge this gap, we propose Style-Extracting Diffusion Models, featuring two conditioning mechanisms. Specifically, we utilize 1) a style conditioning mechanism which allows to inject style information of previously unseen images during image generation and 2) a content conditioning which can be targeted to a downstream task, e.g., layout for segmentation. We introduce a trainable style encoder to extract style information from images, and an aggregation block that merges style information from multiple style inputs. This architecture enables the generation of images with unseen styles in a zero-shot manner, by leveraging styles from unseen images, resulting in more diverse generations. In this work, we use the image layout as target condition and first show the capability of our method on a natural image dataset as a proof-of-concept. We further demonstrate its versatility in histopathology, where we combine prior knowledge about tissue composition and unannotated data to create diverse synthetic images with known layouts. This allows us to generate additional synthetic data to train a segmentation network in a semi-supervised fashion. We verify the added value of the generated images by showing improved segmentation results and lower performance variability between patients when synthetic images are included during segmentation training. Our code will be made publicly available at [LINK].
End-to-end deep learning improves breast cancer classification on
diffusion-weighted MR images (DWI) using a convolutional neural network (CNN)
architecture. A limitation of CNN as opposed to previous model-based approaches
is the dependence on specific DWI input channels used during training. However,
in the context of large-scale application, methods agnostic towards
heterogeneous inputs are desirable, due to the high deviation of scanning
protocols between clinical sites. We propose model-based domain adaptation to
overcome input dependencies and avoid re-training of networks at clinical sites
by restoring training inputs from altered input channels given during
deployment. We demonstrate the method's significant increase in classification
performance and superiority over implicit domain adaptation provided by
training-schemes operating on model-parameters instead of raw DWI images.
16 Apr 2025
Purpose: To present and evaluate Dafne (deep anatomical federated network), a
freely available decentralized, collaborative deep learning system for the
semantic segmentation of radiological images through federated incremental
learning. Materials and Methods: Dafne is free software with a client-server
architecture. The client side is an advanced user interface that applies the
deep learning models stored on the server to the user's data and allows the
user to check and refine the prediction. Incremental learning is then performed
at the client's side and sent back to the server, where it is integrated into
the root model. Dafne was evaluated locally, by assessing the performance gain
across model generations on 38 MRI datasets of the lower legs, and through the
analysis of real-world usage statistics (n = 639 use-cases). Results: Dafne
demonstrated a statistically improvement in the accuracy of semantic
segmentation over time (average increase of the Dice Similarity Coefficient by
0.007 points/generation on the local validation set, p < 0.001). Qualitatively,
the models showed enhanced performance on various radiologic image types,
including those not present in the initial training sets, indicating good model
generalizability. Conclusion: Dafne showed improvement in segmentation quality
over time, demonstrating potential for learning and generalization.
There are no more papers matching your filters at the moment.