Dana-Farber Cancer Institute
Technische Universität DresdenHarvard Medical SchoolUniversity of GenevaCardiff UniversityGerman Cancer Research Center (DKFZ)Dana-Farber Cancer InstituteHelmholtz-Zentrum Dresden-RossendorfBrigham and Women’s HospitalMaastricht University Medical CenterUniversity of Applied Sciences Western Switzerland (HES-SO)University of SherbrookeCentre Hospitalier Universitaire Vaudois (CHUV)The Netherlands Cancer Institute (NKI)
The Image Biomarker Standardisation Initiative (IBSI) aims to improve reproducibility of radiomics studies by standardising the computational process of extracting image biomarkers (features) from images. We have previously established reference values for 169 commonly used features, created a standard radiomics image processing scheme, and developed reporting guidelines for radiomic studies. However, several aspects are not standardised. Here we present a complete version of a reference manual on the use of convolutional filters in radiomics and quantitative image analysis. Filters, such as wavelets or Laplacian of Gaussian filters, play an important part in emphasising specific image characteristics such as edges and blobs. Features derived from filter response maps were found to be poorly reproducible. This reference manual provides definitions for convolutional filters, parameters that should be reported, reference feature values, and tests to verify software compliance with the reference standard.
Meta's LayerSkip accelerates Large Language Model (LLM) inference by enabling accurate early exit and self-speculative decoding within a single model. It introduces a training recipe with non-uniform layer dropout and early exit loss, allowing the model to produce accurate predictions from intermediate layers and to efficiently verify tokens drafted by its own early layers. This approach achieves 1.34x to 2.16x inference speedups across various LLM sizes and tasks with no degradation in final output accuracy.
 Harvard University
Harvard University the University of TokyoPusan National UniversityHarvard Medical School
the University of TokyoPusan National UniversityHarvard Medical School Technical University of MunichDana-Farber Cancer InstituteMass General BrighamBroad Institute of Harvard and MITHelmholtz Munich – German Research Center for Environment and HealthEmory University School of MedicineNational Cancer Center Exploratory Oncology Research & Clinical Trial Center
Technical University of MunichDana-Farber Cancer InstituteMass General BrighamBroad Institute of Harvard and MITHelmholtz Munich – German Research Center for Environment and HealthEmory University School of MedicineNational Cancer Center Exploratory Oncology Research & Clinical Trial CenterThis paper introduces TITAN, a multimodal foundation model that effectively processes whole slide pathology images and text through a three-stage pretraining approach
31 May 2025
RAG-Gym introduces a comprehensive framework for systematically optimizing Retrieval-Augmented Generation (RAG) language agents through prompt engineering, actor tuning, and critic training, achieving improved generalization and robustness compared to outcome-based reinforcement learning. The framework, including the Re2Search agent, integrates process-level supervision to refine information-seeking behaviors and intermediate reasoning steps.
CLAM, a framework developed by researchers at Brigham and Women's Hospital and Harvard Medical School, enables data-efficient and weakly-supervised deep learning for Whole Slide Image analysis in computational pathology. It achieved macro-averaged AUCs up to 0.991 for renal cell carcinoma subtyping and demonstrated robust generalization across diverse datasets, requiring significantly less labeled data than prior methods.
Foundation models (FMs) have shown transformative potential in radiology by
performing diverse, complex tasks across imaging modalities. Here, we developed
CT-FM, a large-scale 3D image-based pre-trained model designed explicitly for
various radiological tasks. CT-FM was pre-trained using 148,000 computed
tomography (CT) scans from the Imaging Data Commons through label-agnostic
contrastive learning. We evaluated CT-FM across four categories of tasks,
namely, whole-body and tumor segmentation, head CT triage, medical image
retrieval, and semantic understanding, showing superior performance against
state-of-the-art models. Beyond quantitative success, CT-FM demonstrated the
ability to cluster regions anatomically and identify similar anatomical and
structural concepts across scans. Furthermore, it remained robust across
test-retest settings and indicated reasonable salient regions attached to its
embeddings. This study demonstrates the value of large-scale medical imaging
foundation models and by open-sourcing the model weights, code, and data, aims
to support more adaptable, reliable, and interpretable AI solutions in
radiology.
 University of Washington
University of Washington University of Cambridge
University of Cambridge Harvard UniversityPusan National University
Harvard UniversityPusan National University University of British ColumbiaHarvard Medical SchoolDana-Farber Cancer InstituteMass General BrighamBroad Institute of Harvard and MITRutgers Robert Wood Johnson Medical SchoolEmory University School of MedicineJohns Hopkins HospitalBeth-Israel Deaconess Medical Center
University of British ColumbiaHarvard Medical SchoolDana-Farber Cancer InstituteMass General BrighamBroad Institute of Harvard and MITRutgers Robert Wood Johnson Medical SchoolEmory University School of MedicineJohns Hopkins HospitalBeth-Israel Deaconess Medical CenterResearchers developed VORTEX, an artificial intelligence framework that predicts 3D spatial gene expression patterns across entire tissue volumes using 3D morphological imaging data and limited 2D spatial transcriptomics. This method successfully mapped complex expression landscapes and tumor microenvironments in various cancer types, overcoming the limitations of traditional 2D spatial transcriptomics.
Harvard Medical School Mohamed bin Zayed University of Artificial IntelligenceDana-Farber Cancer InstituteINSERMBrigham and Women’s HospitalLausanne University Hospital (CHUV)BC Cancer Research InstituteUniversity of Rouen NormandyUniversité de BrestSheikh Shakhbout Medical CityCHU NantesMD Anderson Cancer CenterUniversity Hospital of BrestCentre Hospitalier Universitaire de Poitiers (CHUP)HES-SO Valais-Wallis University of Applied Sciences and Arts Western SwitzerlandUniversity Hospital ZürichCenter Henri BecquerelInstitut de Cancérologie de l’OuestUniversit
de SherbrookeNantes Universit
Mohamed bin Zayed University of Artificial IntelligenceDana-Farber Cancer InstituteINSERMBrigham and Women’s HospitalLausanne University Hospital (CHUV)BC Cancer Research InstituteUniversity of Rouen NormandyUniversité de BrestSheikh Shakhbout Medical CityCHU NantesMD Anderson Cancer CenterUniversity Hospital of BrestCentre Hospitalier Universitaire de Poitiers (CHUP)HES-SO Valais-Wallis University of Applied Sciences and Arts Western SwitzerlandUniversity Hospital ZürichCenter Henri BecquerelInstitut de Cancérologie de l’OuestUniversit
de SherbrookeNantes Universit
 Mohamed bin Zayed University of Artificial IntelligenceDana-Farber Cancer InstituteINSERMBrigham and Women’s HospitalLausanne University Hospital (CHUV)BC Cancer Research InstituteUniversity of Rouen NormandyUniversité de BrestSheikh Shakhbout Medical CityCHU NantesMD Anderson Cancer CenterUniversity Hospital of BrestCentre Hospitalier Universitaire de Poitiers (CHUP)HES-SO Valais-Wallis University of Applied Sciences and Arts Western SwitzerlandUniversity Hospital ZürichCenter Henri BecquerelInstitut de Cancérologie de l’OuestUniversit
de SherbrookeNantes Universit
Mohamed bin Zayed University of Artificial IntelligenceDana-Farber Cancer InstituteINSERMBrigham and Women’s HospitalLausanne University Hospital (CHUV)BC Cancer Research InstituteUniversity of Rouen NormandyUniversité de BrestSheikh Shakhbout Medical CityCHU NantesMD Anderson Cancer CenterUniversity Hospital of BrestCentre Hospitalier Universitaire de Poitiers (CHUP)HES-SO Valais-Wallis University of Applied Sciences and Arts Western SwitzerlandUniversity Hospital ZürichCenter Henri BecquerelInstitut de Cancérologie de l’OuestUniversit
de SherbrookeNantes UniversitWe present a publicly available multimodal dataset for head and neck cancer research, comprising 1123 annotated Positron Emission Tomography/Computed Tomography (PET/CT) studies from patients with histologically confirmed disease, acquired from 10 international medical centers. All studies contain co-registered PET/CT scans with varying acquisition protocols, reflecting real-world clinical diversity from a long-term, multi-institution retrospective collection. Primary gross tumor volumes (GTVp) and involved lymph nodes (GTVn) were manually segmented by experienced radiation oncologists and radiologists following established guidelines. We provide anonymized NifTi files, expert-annotated segmentation masks, comprehensive clinical metadata, and radiotherapy dose distributions for a patient subset. The metadata include TNM staging, HPV status, demographics, long-term follow-up outcomes, survival times, censoring indicators, and treatment information. To demonstrate its utility, we benchmark three key clinical tasks: automated tumor segmentation, recurrence-free survival prediction, and HPV status classification, using state-of-the-art deep learning models like UNet, SegResNet, and multimodal prognostic frameworks.
A general-purpose self-supervised vision model for computational pathology, UNI was trained on a diverse dataset of over 100 million tissue patches, achieving superior performance across 33 clinical tasks and enhanced generalization. The model demonstrated strong data efficiency, outperforming baselines with less labeled data and maintaining performance across different image resolutions.
THREADS is a molecular-driven foundation model for oncologic pathology that learns universal whole-slide image representations by integrating visual features with genomic and transcriptomic data during pretraining. It achieves state-of-the-art performance across 54 diverse oncology tasks, showcasing strong generalizability and data efficiency, and introduces a novel molecular prompting capability for zero-shot-like classification.
 University of Washington
University of Washington Harvard UniversityUniversity of Utah
Harvard UniversityUniversity of Utah Stanford University
Stanford University McGill UniversityHarvard Medical SchoolMassachusetts General HospitalUniversity of Tübingen
McGill UniversityHarvard Medical SchoolMassachusetts General HospitalUniversity of Tübingen The Ohio State UniversityDana-Farber Cancer InstituteAgency for Science Technology and Research (A*STAR)Brigham and Women’s HospitalHelmholtz Center MunichOregon Health & Science UniversityBroad Institute of Harvard and MITHarvard-MITUniversity of Rochester Medical CenterARUP Institute for Clinical and Experimental PathologyBeth-Israel Deaconess Medical Center
The Ohio State UniversityDana-Farber Cancer InstituteAgency for Science Technology and Research (A*STAR)Brigham and Women’s HospitalHelmholtz Center MunichOregon Health & Science UniversityBroad Institute of Harvard and MITHarvard-MITUniversity of Rochester Medical CenterARUP Institute for Clinical and Experimental PathologyBeth-Israel Deaconess Medical CenterKRONOS introduces the first foundation model specifically designed for spatial proteomics, leveraging a massive dataset of 47 million image patches to learn generalizable representations. This model enables superior cell phenotyping, facilitates robust segmentation-free analysis, and improves patient stratification and image retrieval across diverse experimental conditions and tissue types.
27 Sep 2025
Background: Structural variants (SVs) are genomic differences ≥50 bp in length. They remain challenging to detect even with long sequence reads, and the sources of these difficulties are not well quantified.
Results: We identified 35.4 Mb of low-complexity regions (LCRs) in GRCh38. Although these regions cover only 1.2% of the genome, they contain 69.1% of confident SVs in sample HG002. Across long-read SV callers, 77.3-91.3% of erroneous SV calls occur within LCRs, with error rates increasing with LCR length.
Conclusion: SVs are enriched and difficult to call in LCRs. Special care need to be taken for calling and analyzing these variants.
Accurate blind docking has the potential to lead to new biological
breakthroughs, but for this promise to be realized, docking methods must
generalize well across the proteome. Existing benchmarks, however, fail to
rigorously assess generalizability. Therefore, we develop DockGen, a new
benchmark based on the ligand-binding domains of proteins, and we show that
existing machine learning-based docking models have very weak generalization
abilities. We carefully analyze the scaling laws of ML-based docking and show
that, by scaling data and model size, as well as integrating synthetic data
strategies, we are able to significantly increase the generalization capacity
and set new state-of-the-art performance across benchmarks. Further, we propose
Confidence Bootstrapping, a new training paradigm that solely relies on the
interaction between diffusion and confidence models and exploits the
multi-resolution generation process of diffusion models. We demonstrate that
Confidence Bootstrapping significantly improves the ability of ML-based docking
methods to dock to unseen protein classes, edging closer to accurate and
generalizable blind docking methods.
01 Oct 2025
Large language models (LLMs) integrated into agent-driven workflows hold immense promise for healthcare, yet a significant gap exists between their potential and practical implementation within clinical settings. To address this, we present a practitioner-oriented field manual for deploying generative agents that use electronic health record (EHR) data. This guide is informed by our experience deploying the "irAE-Agent", an automated system to detect immune-related adverse events from clinical notes at Mass General Brigham, and by structured interviews with 20 clinicians, engineers, and informatics leaders involved in the project. Our analysis reveals a critical misalignment in clinical AI development: less than 20% of our effort was dedicated to prompt engineering and model development, while over 80% was consumed by the sociotechnical work of implementation. We distill this effort into five "heavy lifts": data integration, model validation, ensuring economic value, managing system drift, and governance. By providing actionable solutions for each of these challenges, this field manual shifts the focus from algorithmic development to the essential infrastructure and implementation work required to bridge the "valley of death" and successfully translate generative AI from pilot projects into routine clinical care.
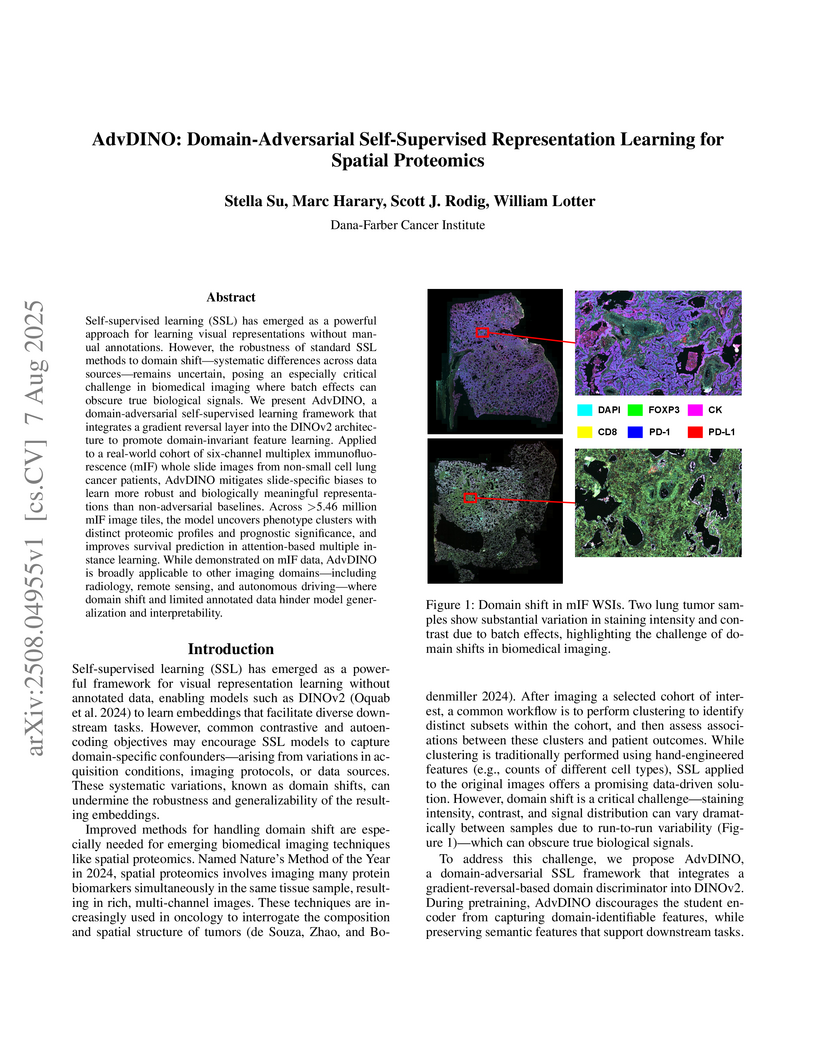
Self-supervised learning (SSL) has emerged as a powerful approach for learning visual representations without manual annotations. However, the robustness of standard SSL methods to domain shift -- systematic differences across data sources -- remains uncertain, posing an especially critical challenge in biomedical imaging where batch effects can obscure true biological signals. We present AdvDINO, a domain-adversarial self-supervised learning framework that integrates a gradient reversal layer into the DINOv2 architecture to promote domain-invariant feature learning. Applied to a real-world cohort of six-channel multiplex immunofluorescence (mIF) whole slide images from non-small cell lung cancer patients, AdvDINO mitigates slide-specific biases to learn more robust and biologically meaningful representations than non-adversarial baselines. Across >5.46 million mIF image tiles, the model uncovers phenotype clusters with distinct proteomic profiles and prognostic significance, and improves survival prediction in attention-based multiple instance learning. While demonstrated on mIF data, AdvDINO is broadly applicable to other imaging domains -- including radiology, remote sensing, and autonomous driving -- where domain shift and limited annotated data hinder model generalization and interpretability.
Nanoparticles (NPs) formed in nonthermal plasmas (NTPs) can have unique properties and applications. However, modeling their growth in these environments presents significant challenges due to the non-equilibrium nature of NTPs, making them computationally expensive to describe. In this work, we address the challenges associated with accelerating the estimation of parameters needed for these models. Specifically, we explore how different machine learning models can be tailored to improve prediction outcomes. We apply these methods to reactive classical molecular dynamics data, which capture the processes associated with colliding silane fragments in NTPs. These reactions exemplify processes where qualitative trends are clear, but their quantification is challenging, hard to generalize, and requires time-consuming simulations. Our results demonstrate that good prediction performance can be achieved when appropriate loss functions are implemented and correct invariances are imposed. While the diversity of molecules used in the training set is critical for accurate prediction, our findings indicate that only a fraction (15-25\%) of the energy and temperature sampling is required to achieve high levels of accuracy. This suggests a substantial reduction in computational effort is possible for similar systems.
Survival models are a popular tool for the analysis of time to event data with applications in medicine, engineering, economics, and many more. Advances like the Cox proportional hazard model have enabled researchers to better describe hazard rates for the occurrence of single fatal events, but are unable to accurately model competing events and transitions. Common phenomena are often better described through multiple states, for example: the progress of a disease modeled as healthy, sick and dead instead of healthy and dead, where the competing nature of death and disease has to be taken into account. Moreover, Cox models are limited by modeling assumptions, like proportionality of hazard rates and linear effects. Individual characteristics can vary significantly between observational units, like patients, resulting in idiosyncratic hazard rates and different disease trajectories. These considerations require flexible modeling assumptions. To overcome these issues, we propose the use of neural ordinary differential equations as a flexible and general method for estimating multi-state survival models by directly solving the Kolmogorov forward equations. To quantify the uncertainty in the resulting individual cause-specific hazard rates, we further introduce a variational latent variable model and show that this enables meaningful clustering with respect to multi-state outcomes as well as interpretability regarding covariate values. We show that our model exhibits state-of-the-art performance on popular survival data sets and demonstrate its efficacy in a multi-state setting
Clinical notes contain rich data, which is unexploited in predictive modeling
compared to structured data. In this work, we developed a new text
representation Clinical XLNet for clinical notes which also leverages the
temporal information of the sequence of the notes. We evaluated our models on
prolonged mechanical ventilation prediction problem and our experiments
demonstrated that Clinical XLNet outperforms the best baselines consistently.
Researchers at Brigham and Women’s Hospital, Harvard Medical School, and the Broad Institute developed Pathomic Fusion, an integrated deep learning framework for combining histopathology images and genomic profiles to improve cancer diagnosis and prognosis. The framework achieved a c-Index of 0.826 for glioma survival prediction, a 6.31% improvement over prior deep learning methods, and provides interpretable insights into both morphological and molecular features influencing outcomes.
Graph Neural Networks (GNNs) traditionally employ a message-passing mechanism
that resembles diffusion over undirected graphs, which often leads to
homogenization of node features and reduced discriminative power in tasks such
as node classification. Our key insight for addressing this limitation is to
assign fuzzy edge directions -- that can vary continuously from node i
pointing to node j to vice versa -- to the edges of a graph so that features
can preferentially flow in one direction between nodes to enable long-range
information transmission across the graph. We also introduce a novel
complex-valued Laplacian for directed graphs with fuzzy edges where the real
and imaginary parts represent information flow in opposite directions. Using
this Laplacian, we propose a general framework, called Continuous Edge
Direction (CoED) GNN, for learning on graphs with fuzzy edges and prove its
expressivity limits using a generalization of the Weisfeiler-Leman (WL) graph
isomorphism test for directed graphs with fuzzy edges. Our architecture
aggregates neighbor features scaled by the learned edge directions and
processes the aggregated messages from in-neighbors and out-neighbors
separately alongside the self-features of the nodes. Since continuous edge
directions are differentiable, they can be learned jointly with the GNN weights
via gradient-based optimization. CoED GNN is particularly well-suited for graph
ensemble data where the graph structure remains fixed but multiple realizations
of node features are available, such as in gene regulatory networks, web
connectivity graphs, and power grids. We demonstrate through extensive
experiments on both synthetic and real graph ensemble datasets that learning
continuous edge directions significantly improves performance both for
undirected and directed graphs compared with existing methods.
There are no more papers matching your filters at the moment.