Erasmus MC
The global navigation satellite systems (GNSS) play a vital role in transport
systems for accurate and consistent vehicle localization. However, GNSS
observations can be distorted due to multipath effects and non-line-of-sight
(NLOS) receptions in challenging environments such as urban canyons. In such
cases, traditional methods to classify and exclude faulty GNSS observations may
fail, leading to unreliable state estimation and unsafe system operations. This
work proposes a deep-learning-based method to detect NLOS receptions and
predict GNSS pseudorange errors by analyzing GNSS observations as a
spatio-temporal modeling problem. Compared to previous works, we construct a
transformer-like attention mechanism to enhance the long short-term memory
(LSTM) networks, improving model performance and generalization. For the
training and evaluation of the proposed network, we used labeled datasets from
the cities of Hong Kong and Aachen. We also introduce a dataset generation
process to label the GNSS observations using lidar maps. In experimental
studies, we compare the proposed network with a deep-learning-based model and
classical machine-learning models. Furthermore, we conduct ablation studies of
our network components and integrate the NLOS detection with data
out-of-distribution in a state estimator. As a result, our network presents
improved precision and recall ratios compared to other models. Additionally, we
show that the proposed method avoids trajectory divergence in real-world
vehicle localization by classifying and excluding NLOS observations.
Improving Patient Pre-screening for Clinical Trials: Assisting
Physicians with Large Language Models
Improving Patient Pre-screening for Clinical Trials: Assisting
Physicians with Large Language Models
Physicians considering clinical trials for their patients are met with the
laborious process of checking many text based eligibility criteria. Large
Language Models (LLMs) have shown to perform well for clinical information
extraction and clinical reasoning, including medical tests, but not yet in
real-world scenarios. This paper investigates the use of InstructGPT to assist
physicians in determining eligibility for clinical trials based on a patient's
summarised medical profile. Using a prompting strategy combining one-shot,
selection-inference and chain-of-thought techniques, we investigate the
performance of LLMs on 10 synthetically created patient profiles. Performance
is evaluated at four levels: ability to identify screenable eligibility
criteria from a trial given a medical profile; ability to classify for each
individual criterion whether the patient qualifies; the overall classification
whether a patient is eligible for a clinical trial and the percentage of
criteria to be screened by physician. We evaluated against 146 clinical trials
and a total of 4,135 eligibility criteria. The LLM was able to correctly
identify the screenability of 72% (2,994/4,135) of the criteria. Additionally,
72% (341/471) of the screenable criteria were evaluated correctly. The
resulting trial level classification as eligible or ineligible resulted in a
recall of 0.5. By leveraging LLMs with a physician-in-the-loop, a recall of 1.0
and precision of 0.71 on clinical trial level can be achieved while reducing
the amount of criteria to be checked by an estimated 90%. LLMs can be used to
assist physicians with pre-screening of patients for clinical trials. By
forcing instruction-tuned LLMs to produce chain-of-thought responses, the
reasoning can be made transparent to and the decision process becomes amenable
by physicians, thereby making such a system feasible for use in real-world
scenarios.
Synthetic Aperture Radar (SAR) data enables large-scale surveillance of maritime vessels. However, near-real-time monitoring is currently constrained by the need to downlink all raw data, perform image focusing, and subsequently analyze it on the ground. On-board processing to generate higher-level products could reduce the data volume that needs to be downlinked, alleviating bandwidth constraints and minimizing latency. However, traditional image focusing and processing algorithms face challenges due to the satellite's limited memory, processing power, and computational resources. This work proposes and evaluates neural networks designed for real-time inference on unfocused SAR data acquired in Stripmap and Interferometric Wide (IW) modes captured with Sentinel-1. Our results demonstrate the feasibility of using one of our models for on-board processing and deployment on an FPGA. Additionally, by investigating a binary classification task between ships and windmills, we demonstrate that target classification is possible.
We propose to apply non-linear representation learning to voxelwise rs-fMRI
data. Learning the non-linear representations is done using a variational
autoencoder (VAE). The VAE is trained on voxelwise rs-fMRI data and performs
non-linear dimensionality reduction that retains meaningful information. The
retention of information in the model's representations is evaluated using
downstream age regression and sex classification tasks. The results on these
tasks are highly encouraging and a linear regressor trained with the
representations of our unsupervised model performs almost as well as a
supervised neural network, trained specifically for age regression on the same
dataset. The model is also evaluated with a schizophrenia diagnosis prediction
task, to assess its feasibility as a dimensionality reduction method for
neuropsychiatric datasets. These results highlight the potential for
pre-training on a larger set of individuals who do not have mental illness, to
improve the downstream neuropsychiatric task results. The pre-trained model is
fine-tuned for a variable number of epochs on a schizophrenia dataset and we
find that fine-tuning for 1 epoch yields the best results. This work therefore
not only opens up non-linear dimensionality reduction for voxelwise rs-fMRI
data but also shows that pre-training a deep learning model on voxelwise
rs-fMRI datasets greatly increases performance even on smaller datasets. It
also opens up the ability to look at the distribution of rs-fMRI time series in
the latent space of the VAE for heterogeneous neuropsychiatric disorders like
schizophrenia in future work. This can be complemented with the generative
aspect of the model that allows us to reconstruct points from the model's
latent space back into brain space and obtain an improved understanding of the
relation that the VAE learns between subjects, timepoints, and a subject's
characteristics.
The scarcity of labeled data often limits the application of supervised deep learning techniques for medical image segmentation. This has motivated the development of semi-supervised techniques that learn from a mixture of labeled and unlabeled images. In this paper, we propose a novel semi-supervised method that, in addition to supervised learning on labeled training images, learns to predict segmentations consistent under a given class of transformations on both labeled and unlabeled images. More specifically, in this work we explore learning equivariance to elastic deformations. We implement this through: 1) a Siamese architecture with two identical branches, each of which receives a differently transformed image, and 2) a composite loss function with a supervised segmentation loss term and an unsupervised term that encourages segmentation consistency between the predictions of the two branches. We evaluate the method on a public dataset of chest radiographs with segmentations of anatomical structures using 5-fold cross-validation. The proposed method reaches significantly higher segmentation accuracy compared to supervised learning. This is due to learning transformation consistency on both labeled and unlabeled images, with the latter contributing the most. We achieve the performance comparable to state-of-the-art chest X-ray segmentation methods while using substantially fewer labeled images.
The event-based model (EBM) for data-driven disease progression modeling estimates the sequence in which biomarkers for a disease become abnormal. This helps in understanding the dynamics of disease progression and facilitates early diagnosis by staging patients on a disease progression timeline. Existing EBM methods are all generative in nature. In this work we propose a novel discriminative approach to EBM, which is shown to be more accurate as well as computationally more efficient than existing state-of-the art EBM methods. The method first estimates for each subject an approximate ordering of events, by ranking the posterior probabilities of individual biomarkers being abnormal. Subsequently, the central ordering over all subjects is estimated by fitting a generalized Mallows model to these approximate subject-specific orderings based on a novel probabilistic Kendall's Tau distance. To evaluate the accuracy, we performed extensive experiments on synthetic data simulating the progression of Alzheimer's disease. Subsequently, the method was applied to the Alzheimer's Disease Neuroimaging Initiative (ADNI) data to estimate the central event ordering in the dataset. The experiments benchmark the accuracy of the new model under various conditions and compare it with existing state-of-the-art EBM methods. The results indicate that discriminative EBM could be a simple and elegant approach to disease progression modeling.
 University of Cambridge
University of Cambridge University of Southern California
University of Southern California National University of Singapore
National University of Singapore University College London
University College London University of Oxford
University of Oxford Georgia Institute of Technology
Georgia Institute of Technology University of Copenhagen
University of Copenhagen University of California, San Diego
University of California, San Diego McGill University
McGill University Emory University
Emory University University of Pennsylvania
University of Pennsylvania Arizona State University
Arizona State University University of Maryland
University of Maryland King’s College LondonErasmus MCMayo ClinicLund UniversityBrandeis UniversityBen-Gurion University of the NegevUniversity of Eastern FinlandPortland State UniversityUniversity of California San FranciscoNational Institute on AgingGenentechUniversity of PlymouthBanner Alzheimer’s InstituteUCL Queen Square Institute of NeurologyThe University of Texas Health Science Center at HoustonMedical College of WisconsinGerman Center for Neurodegenerative DiseasesUniversity of GhanaInstituto Tecnol ́ogico y de Estudios Superiores de MonterreyH. Lundbeck A/SVU Medical CentreInstitut du Cerveau et de la Moelle ́epini`ereVasile Lucaciu National CollegeBiomarinIBM Research - Australia
King’s College LondonErasmus MCMayo ClinicLund UniversityBrandeis UniversityBen-Gurion University of the NegevUniversity of Eastern FinlandPortland State UniversityUniversity of California San FranciscoNational Institute on AgingGenentechUniversity of PlymouthBanner Alzheimer’s InstituteUCL Queen Square Institute of NeurologyThe University of Texas Health Science Center at HoustonMedical College of WisconsinGerman Center for Neurodegenerative DiseasesUniversity of GhanaInstituto Tecnol ́ogico y de Estudios Superiores de MonterreyH. Lundbeck A/SVU Medical CentreInstitut du Cerveau et de la Moelle ́epini`ereVasile Lucaciu National CollegeBiomarinIBM Research - AustraliaWe present the findings of "The Alzheimer's Disease Prediction Of
Longitudinal Evolution" (TADPOLE) Challenge, which compared the performance of
92 algorithms from 33 international teams at predicting the future trajectory
of 219 individuals at risk of Alzheimer's disease. Challenge participants were
required to make a prediction, for each month of a 5-year future time period,
of three key outcomes: clinical diagnosis, Alzheimer's Disease Assessment Scale
Cognitive Subdomain (ADAS-Cog13), and total volume of the ventricles. The
methods used by challenge participants included multivariate linear regression,
machine learning methods such as support vector machines and deep neural
networks, as well as disease progression models. No single submission was best
at predicting all three outcomes. For clinical diagnosis and ventricle volume
prediction, the best algorithms strongly outperform simple baselines in
predictive ability. However, for ADAS-Cog13 no single submitted prediction
method was significantly better than random guesswork. Two ensemble methods
based on taking the mean and median over all predictions, obtained top scores
on almost all tasks. Better than average performance at diagnosis prediction
was generally associated with the additional inclusion of features from
cerebrospinal fluid (CSF) samples and diffusion tensor imaging (DTI). On the
other hand, better performance at ventricle volume prediction was associated
with inclusion of summary statistics, such as the slope or maxima/minima of
biomarkers. TADPOLE's unique results suggest that current prediction algorithms
provide sufficient accuracy to exploit biomarkers related to clinical diagnosis
and ventricle volume, for cohort refinement in clinical trials for Alzheimer's
disease. However, results call into question the usage of cognitive test scores
for patient selection and as a primary endpoint in clinical trials.
Segmentations are crucial in medical imaging to obtain morphological, volumetric, and radiomics biomarkers. Manual segmentation is accurate but not feasible in the radiologist's clinical workflow, while automatic segmentation generally obtains sub-par performance. We therefore developed a minimally interactive deep learning-based segmentation method for soft-tissue tumors (STTs) on CT and MRI. The method requires the user to click six points near the tumor's extreme boundaries. These six points are transformed into a distance map and serve, with the image, as input for a Convolutional Neural Network. For training and validation, a multicenter dataset containing 514 patients and nine STT types in seven anatomical locations was used, resulting in a Dice Similarity Coefficient (DSC) of 0.85±0.11 (mean ± standard deviation (SD)) for CT and 0.84±0.12 for T1-weighted MRI, when compared to manual segmentations made by expert radiologists. Next, the method was externally validated on a dataset including five unseen STT phenotypes in extremities, achieving 0.81±0.08 for CT, 0.84±0.09 for T1-weighted MRI, and 0.88\pm0.08 for previously unseen T2-weighted fat-saturated (FS) MRI. In conclusion, our minimally interactive segmentation method effectively segments different types of STTs on CT and MRI, with robust generalization to previously unseen phenotypes and imaging modalities.
Augmenting X-ray imaging with 3D roadmap to improve guidance is a common
strategy. Such approaches benefit from automated analysis of the X-ray images,
such as the automatic detection and tracking of instruments. In this paper, we
propose a real-time method to segment the catheter and guidewire in 2D X-ray
fluoroscopic sequences. The method is based on deep convolutional neural
networks. The network takes as input the current image and the three previous
ones, and segments the catheter and guidewire in the current image.
Subsequently, a centerline model of the catheter is constructed from the
segmented image. A small set of annotated data combined with data augmentation
is used to train the network. We trained the method on images from 182 X-ray
sequences from 23 different interventions. On a testing set with images of 55
X-ray sequences from 5 other interventions, a median centerline distance error
of 0.2 mm and a median tip distance error of 0.9 mm was obtained. The
segmentation of the instruments in 2D X-ray sequences is performed in a
real-time fully-automatic manner.
Tract-specific diffusion measures, as derived from brain diffusion MRI, have been linked to white matter tract structural integrity and neurodegeneration. As a consequence, there is a large interest in the automatic segmentation of white matter tract in diffusion tensor MRI data. Methods based on the tractography are popular for white matter tract segmentation. However, because of the limited consistency and long processing time, such methods may not be suitable for clinical practice. We therefore developed a novel convolutional neural network based method to directly segment white matter tract trained on a low-resolution dataset of 9149 DTI images. The method is optimized on input, loss function and network architecture selections. We evaluated both segmentation accuracy and reproducibility, and reproducibility of determining tract-specific diffusion measures. The reproducibility of the method is higher than that of the reference standard and the determined diffusion measures are consistent. Therefore, we expect our method to be applicable in clinical practice and in longitudinal analysis of white matter microstructure.
Computational Pathology (CPATH) systems have the potential to automate
diagnostic tasks. However, the artifacts on the digitized histological glass
slides, known as Whole Slide Images (WSIs), may hamper the overall performance
of CPATH systems. Deep Learning (DL) models such as Vision Transformers (ViTs)
may detect and exclude artifacts before running the diagnostic algorithm. A
simple way to develop robust and generalized ViTs is to train them on massive
datasets. Unfortunately, acquiring large medical datasets is expensive and
inconvenient, prompting the need for a generalized artifact detection method
for WSIs. In this paper, we present a student-teacher recipe to improve the
classification performance of ViT for the air bubbles detection task. ViT,
trained under the student-teacher framework, boosts its performance by
distilling existing knowledge from the high-capacity teacher model. Our
best-performing ViT yields 0.961 and 0.911 F1-score and MCC, respectively,
observing a 7% gain in MCC against stand-alone training. The proposed method
presents a new perspective of leveraging knowledge distillation over transfer
learning to encourage the use of customized transformers for efficient
preprocessing pipelines in the CPATH systems.
Standardized alignment of the embryo in three-dimensional (3D) ultrasound images aids prenatal growth monitoring by facilitating standard plane detection, improving visualization of landmarks and accentuating differences between different scans. In this work, we propose an automated method for standardizing this alignment. Given a segmentation mask of the embryo, Principal Component Analysis (PCA) is applied to the mask extracting the embryo's principal axes, from which four candidate orientations are derived. The candidate in standard orientation is selected using one of three strategies: a heuristic based on Pearson's correlation assessing shape, image matching to an atlas through normalized cross-correlation, and a Random Forest classifier. We tested our method on 2166 images longitudinally acquired 3D ultrasound scans from 1043 pregnancies from the Rotterdam Periconceptional Cohort, ranging from 7+0 to 12+6 weeks of gestational age. In 99.0% of images, PCA correctly extracted the principal axes of the embryo. The correct candidate was selected by the Pearson Heuristic, Atlas-based and Random Forest in 97.4%, 95.8%, and 98.4% of images, respectively. A Majority Vote of these selection methods resulted in an accuracy of 98.5%. The high accuracy of this pipeline enables consistent embryonic alignment in the first trimester, enabling scalable analysis in both clinical and research settings. The code is publicly available at: this https URL.
While foundation models (FMs) offer strong potential for AI-based dementia diagnosis, their integration into federated learning (FL) systems remains underexplored. In this benchmarking study, we systematically evaluate the impact of key design choices: classification head architecture, fine-tuning strategy, and aggregation method, on the performance and efficiency of federated FM tuning using brain MRI data. Using a large multi-cohort dataset, we find that the architecture of the classification head substantially influences performance, freezing the FM encoder achieves comparable results to full fine-tuning, and advanced aggregation methods outperform standard federated averaging. Our results offer practical insights for deploying FMs in decentralized clinical settings and highlight trade-offs that should guide future method development.
Advances in additive manufacturing have enabled the realisation of inexpensive, scalable, diffractive acoustic lenses that can be used to generate complex acoustic fields via phase and/or amplitude modulation. However, the design of these holograms relies on a thin-element approximation adapted from optics which can severely limit the fidelity of the realised acoustic field. Here, we introduce physics-based acoustic holograms with a complex internal structure. The structures are designed using a differentiable acoustic model with manufacturing constraints via optimisation of the acoustic property distribution within the hologram. The holograms can be fabricated simply and inexpensively using contemporary 3D printers. Experimental measurements demonstrate a significant improvement compared to conventional thin-element holograms.
Cardiac magnetic resonance (CMR) is used extensively in the diagnosis and
management of cardiovascular disease. Deep learning methods have proven to
deliver segmentation results comparable to human experts in CMR imaging, but
there have been no convincing results for the problem of end-to-end
segmentation and diagnosis from CMR. This is in part due to a lack of
sufficiently large datasets required to train robust diagnosis models. In this
paper, we propose a learning method to train diagnosis models, where our
approach is designed to work with relatively small datasets. In particular, the
optimisation loss is based on multi-task learning that jointly trains for the
tasks of segmentation and diagnosis classification. We hypothesize that
segmentation has a regularizing effect on the learning of features relevant for
diagnosis. Using the 100 training and 50 testing samples available from the
Automated Cardiac Diagnosis Challenge (ACDC) dataset, which has a balanced
distribution of 5 cardiac diagnoses, we observe a reduction of the
classification error from 32% to 22%, and a faster convergence compared to a
baseline without segmentation. To the best of our knowledge, this is the best
diagnosis results from CMR using an end-to-end diagnosis and segmentation
learning method.
01 Aug 2024
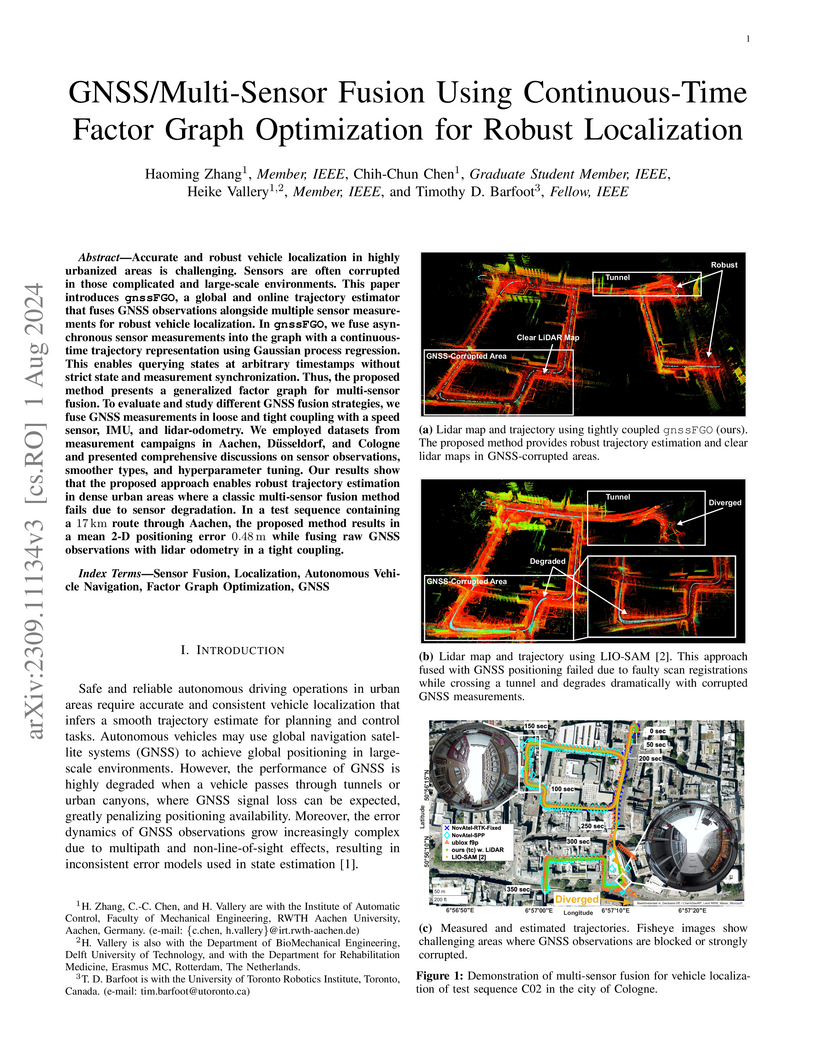
Accurate and robust vehicle localization in highly urbanized areas is challenging. Sensors are often corrupted in those complicated and large-scale environments. This paper introduces GNSS-FGO, an online and global trajectory estimator that fuses GNSS observations alongside multiple sensor measurements for robust vehicle localization. In GNSS-FGO, we fuse asynchronous sensor measurements into the graph with a continuous-time trajectory representation using Gaussian process regression. This enables querying states at arbitrary timestamps so that sensor observations are fused without requiring strict state and measurement synchronization. Thus, the proposed method presents a generalized factor graph for multi-sensor fusion. To evaluate and study different GNSS fusion strategies, we fuse GNSS measurements in loose and tight coupling with a speed sensor, IMU, and lidar-odometry. We employed datasets from measurement campaigns in Aachen, Duesseldorf, and Cologne in experimental studies and presented comprehensive discussions on sensor observations, smoother types, and hyperparameter tuning. Our results show that the proposed approach enables robust trajectory estimation in dense urban areas, where the classic multi-sensor fusion method fails due to sensor degradation. In a test sequence containing a 17km route through Aachen, the proposed method results in a mean 2D positioning error of 0.48m while fusing raw GNSS observations with lidar odometry in a tight coupling.
In medical-data driven learning, 3D convolutional neural networks (CNNs) have
started to show superior performance to 2D CNNs in numerous deep learning
tasks, proving the added value of 3D spatial information in feature
representation. However, the difficulty in collecting more training samples to
converge, more computational resources and longer execution time make this
approach less applied. Also, applying transfer learning on 3D CNN is
challenging due to a lack of publicly available pre-trained 3D models. To
tackle these issues, we proposed a novel 2D strategical representation of
volumetric data, namely 2.75D. In this work, the spatial information of 3D
images is captured in a single 2D view by a spiral-spinning technique. As a
result, 2D CNN networks can also be used to learn volumetric information.
Besides, we can fully leverage pre-trained 2D CNNs for downstream vision
problems. We also explore a multi-view 2.75D strategy, 2.75D 3 channels
(2.75Dx3), to boost the advantage of 2.75D. We evaluated the proposed methods
on three public datasets with different modalities or organs (Lung CT, Breast
MRI, and Prostate MRI), against their 2D, 2.5D, and 3D counterparts in
classification tasks. Results show that the proposed methods significantly
outperform other counterparts when all methods were trained from scratch on the
lung dataset. Such performance gain is more pronounced with transfer learning
or in the case of limited training data. Our methods also achieved comparable
performance on other datasets. In addition, our methods achieved a substantial
reduction in time consumption of training and inference compared with the 2.5D
or 3D method.
In this paper, we present a new approach for uncertainty-aware retinal layer segmentation in Optical Coherence Tomography (OCT) scans using probabilistic signed distance functions (SDF). Traditional pixel-wise and regression-based methods primarily encounter difficulties in precise segmentation and lack of geometrical grounding respectively. To address these shortcomings, our methodology refines the segmentation by predicting a signed distance function (SDF) that effectively parameterizes the retinal layer shape via level set. We further enhance the framework by integrating probabilistic modeling, applying Gaussian distributions to encapsulate the uncertainty in the shape parameterization. This ensures a robust representation of the retinal layer morphology even in the presence of ambiguous input, imaging noise, and unreliable segmentations. Both quantitative and qualitative evaluations demonstrate superior performance when compared to other methods. Additionally, we conducted experiments on artificially distorted datasets with various noise types-shadowing, blinking, speckle, and motion-common in OCT scans to showcase the effectiveness of our uncertainty estimation. Our findings demonstrate the possibility to obtain reliable segmentation of retinal layers, as well as an initial step towards the characterization of layer integrity, a key biomarker for disease progression. Our code is available at \url{this https URL}.
The purposes of this study were to investigate: 1) the effect of placement of
region-of-interest (ROI) for texture analysis of subchondral bone in knee
radiographs, and 2) the ability of several texture descriptors to distinguish
between the knees with and without radiographic osteoarthritis (OA). Bilateral
posterior-anterior knee radiographs were analyzed from the baseline of OAI and
MOST datasets. A fully automatic method to locate the most informative region
from subchondral bone using adaptive segmentation was developed. We used an
oversegmentation strategy for partitioning knee images into the compact regions
that follow natural texture boundaries. LBP, Fractal Dimension (FD), Haralick
features, Shannon entropy, and HOG methods were computed within the standard
ROI and within the proposed adaptive ROIs. Subsequently, we built logistic
regression models to identify and compare the performances of each texture
descriptor and each ROI placement method using 5-fold cross validation setting.
Importantly, we also investigated the generalizability of our approach by
training the models on OAI and testing them on MOST dataset.We used area under
the receiver operating characteristic (ROC) curve (AUC) and average precision
(AP) obtained from the precision-recall (PR) curve to compare the results. We
found that the adaptive ROI improves the classification performance (OA vs.
non-OA) over the commonly used standard ROI (up to 9% percent increase in AUC).
We also observed that, from all texture parameters, LBP yielded the best
performance in all settings with the best AUC of 0.840 [0.825, 0.852] and
associated AP of 0.804 [0.786, 0.820]. Compared to the current state-of-the-art
approaches, our results suggest that the proposed adaptive ROI approach in
texture analysis of subchondral bone can increase the diagnostic performance
for detecting the presence of radiographic OA.
Seizures occur in the neonatal period more frequently than other periods of
life and usually denote the presence of serious brain dysfunction. The gold
standard for detecting seizures is based on visual inspection of continuous
electroencephalogram (cEEG) complemented by video analysis, performed by an
expert clinical neurophysiologist. Previous studies have reported varying
degree of agreement between expert EEG readers, with kappa coefficients ranging
from 0.4 to 0.85, calling into question the validity of visual scoring. This
variability in visual scoring of neonatal seizures may be due to factors such
as reader expertise and the nature of expressed patterns. One of the possible
reasons for low inter-rater agreement is the absence of any benchmark for the
EEG readers to be able to compare their opinions. One way to develop this is to
use a shared multi-center neonatal seizure database and use the inputs from
multiple experts. This will also improve the teaching of trainees, and help to
avoid potential bias from a single expert's opinion. In this paper, we
introduce and explain the NeoGuard public learning platform that can be used by
trainees, tutors, and expert EEG readers who are interested to test their
knowledge and learn from neonatal EEG-polygraphic segments scored by several
expert EEG readers. For this platform, 1919 clinically relevant segments,
totaling 280h, recorded from 71 term neonates in two centers, including a wide
variety of seizures and artifacts were used. These segments were scored by 4
EEG readers from three different centers. Users of this platform can score an
arbitrary number of segments and then test their scoring with the experts'
opinions. The kappa and joint probability of agreement, is then shown as
inter-rater agreement metrics between the user and each of the experts. The
platform is publicly available at the NeoGuard website (www.neoguard.net).
There are no more papers matching your filters at the moment.