IQVIA
12 Mar 2025
A comprehensive survey examines the current state, challenges, and future directions of agentic AI systems in scientific discovery, analyzing existing frameworks like LitSearch and ResearchArena while providing a systematic categorization of tools, datasets, and evaluation metrics used across chemistry, biology, and materials science applications.
Clinical trials are pivotal for developing new medical treatments but typically carry risks such as patient mortality and enrollment failure that waste immense efforts spanning over a decade. Applying artificial intelligence (AI) to predict key events in clinical trials holds great potential for providing insights to guide trial designs. However, complex data collection and question definition requiring medical expertise have hindered the involvement of AI thus far. This paper tackles these challenges by presenting a comprehensive suite of 23 meticulously curated AI-ready datasets covering multi-modal input features and 8 crucial prediction challenges in clinical trial design, encompassing prediction of trial duration, patient dropout rate, serious adverse event, mortality rate, trial approval outcome, trial failure reason, drug dose finding, design of eligibility criteria. Furthermore, we provide basic validation methods for each task to ensure the datasets' usability and reliability. We anticipate that the availability of such open-access datasets will catalyze the development of advanced AI approaches for clinical trial design, ultimately advancing clinical trial research and accelerating medical solution development.
26 Feb 2025
OntologyRAG, developed by IQVIA's Applied AI Science division, improves biomedical code mapping by integrating large language models with structured ontology knowledge graphs through a retrieval-augmented generation approach. The system accurately assesses semantic relationships between codes and provides transparent reasoning, achieving high precision in identifying clear matches and non-matches across various biomedical classification systems.
Medication recommendation is an important healthcare application. It is commonly formulated as a temporal prediction task. Hence, most existing works only utilize longitudinal electronic health records (EHRs) from a small number of patients with multiple visits ignoring a large number of patients with a single visit (selection bias). Moreover, important hierarchical knowledge such as diagnosis hierarchy is not leveraged in the representation learning process. To address these challenges, we propose G-BERT, a new model to combine the power of Graph Neural Networks (GNNs) and BERT (Bidirectional Encoder Representations from Transformers) for medical code representation and medication recommendation. We use GNNs to represent the internal hierarchical structures of medical codes. Then we integrate the GNN representation into a transformer-based visit encoder and pre-train it on EHR data from patients only with a single visit. The pre-trained visit encoder and representation are then fine-tuned for downstream predictive tasks on longitudinal EHRs from patients with multiple visits. G-BERT is the first to bring the language model pre-training schema into the healthcare domain and it achieved state-of-the-art performance on the medication recommendation task.
Too Late to Train, Too Early To Use? A Study on Necessity and Viability of Low-Resource Bengali LLMs
Too Late to Train, Too Early To Use? A Study on Necessity and Viability of Low-Resource Bengali LLMs
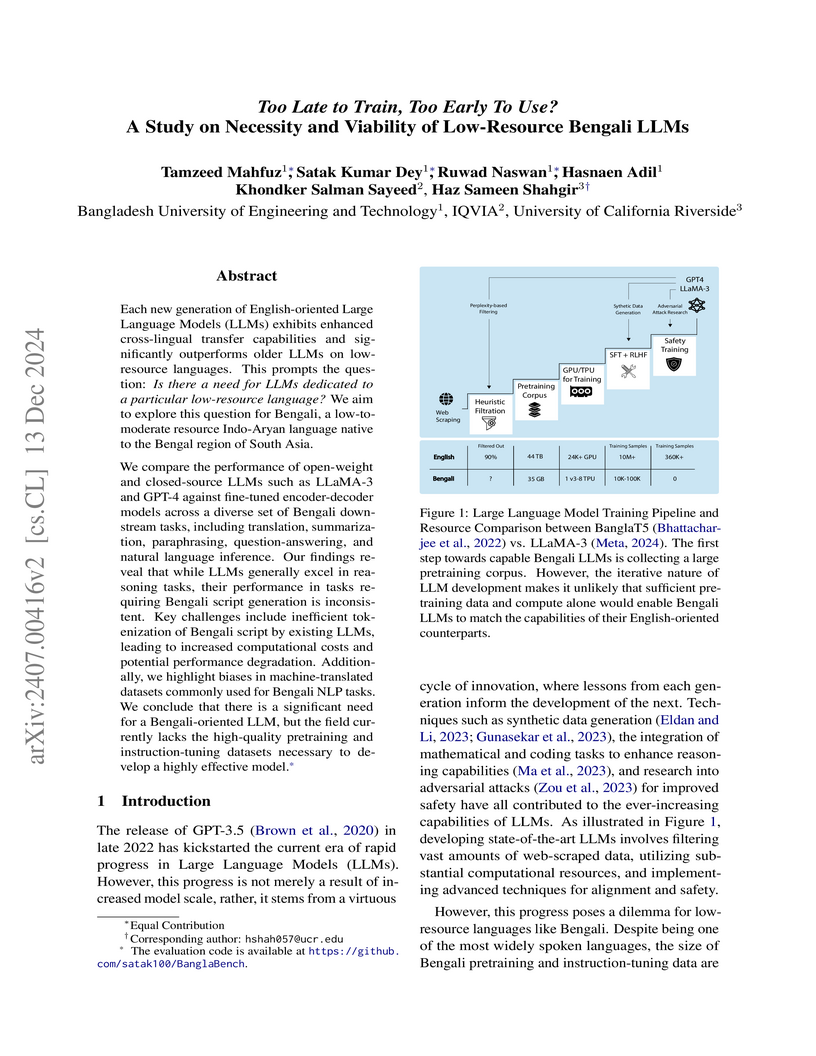
Each new generation of English-oriented Large Language Models (LLMs) exhibits
enhanced cross-lingual transfer capabilities and significantly outperforms
older LLMs on low-resource languages. This prompts the question: Is there a
need for LLMs dedicated to a particular low-resource language? We aim to
explore this question for Bengali, a low-to-moderate resource Indo-Aryan
language native to the Bengal region of South Asia.
We compare the performance of open-weight and closed-source LLMs such as
LLaMA-3 and GPT-4 against fine-tuned encoder-decoder models across a diverse
set of Bengali downstream tasks, including translation, summarization,
paraphrasing, question-answering, and natural language inference. Our findings
reveal that while LLMs generally excel in reasoning tasks, their performance in
tasks requiring Bengali script generation is inconsistent. Key challenges
include inefficient tokenization of Bengali script by existing LLMs, leading to
increased computational costs and potential performance degradation.
Additionally, we highlight biases in machine-translated datasets commonly used
for Bengali NLP tasks. We conclude that there is a significant need for a
Bengali-oriented LLM, but the field currently lacks the high-quality
pretraining and instruction-tuning datasets necessary to develop a highly
effective model.
Taking advantage of the widespread use of ontologies to organise and harmonize knowledge across several distinct domains, this paper proposes a novel approach to improve an embedding-Large Language Model (embedding-LLM) of interest by infusing the knowledge formalized by a reference ontology: ontological knowledge infusion aims at boosting the ability of the considered LLM to effectively model the knowledge domain described by the infused ontology. The linguistic information (i.e. concept synonyms and descriptions) and structural information (i.e. is-a relations) formalized by the ontology are utilized to compile a comprehensive set of concept definitions, with the assistance of a powerful generative LLM (i.e. GPT-3.5-turbo). These concept definitions are then employed to fine-tune the target embedding-LLM using a contrastive learning framework. To demonstrate and evaluate the proposed approach, we utilize the biomedical disease ontology MONDO. The results show that embedding-LLMs enhanced by ontological disease knowledge exhibit an improved capability to effectively evaluate the similarity of in-domain sentences from biomedical documents mentioning diseases, without compromising their out-of-domain performance.
Medication recommendation is an essential task of AI for healthcare. Existing works focused on recommending drug combinations for patients with complex health conditions solely based on their electronic health records. Thus, they have the following limitations: (1) some important data such as drug molecule structures have not been utilized in the recommendation process. (2) drug-drug interactions (DDI) are modeled implicitly, which can lead to sub-optimal results. To address these limitations, we propose a DDI-controllable drug recommendation model named SafeDrug to leverage drugs' molecule structures and model DDIs explicitly. SafeDrug is equipped with a global message passing neural network (MPNN) module and a local bipartite learning module to fully encode the connectivity and functionality of drug molecules. SafeDrug also has a controllable loss function to control DDI levels in the recommended drug combinations effectively. On a benchmark dataset, our SafeDrug is relatively shown to reduce DDI by 19.43% and improves 2.88% on Jaccard similarity between recommended and actually prescribed drug combinations over previous approaches. Moreover, SafeDrug also requires much fewer parameters than previous deep learning-based approaches, leading to faster training by about 14% and around 2x speed-up in inference.
Clinical trials are crucial for drug development but are time consuming, expensive, and often burdensome on patients. More importantly, clinical trials face uncertain outcomes due to issues with efficacy, safety, or problems with patient recruitment. If we were better at predicting the results of clinical trials, we could avoid having to run trials that will inevitably fail more resources could be devoted to trials that are likely to succeed. In this paper, we propose Hierarchical INteraction Network (HINT) for more general, clinical trial outcome predictions for all diseases based on a comprehensive and diverse set of web data including molecule information of the drugs, target disease information, trial protocol and biomedical knowledge. HINT first encode these multi-modal data into latent embeddings, where an imputation module is designed to handle missing data. Next, these embeddings will be fed into the knowledge embedding module to generate knowledge embeddings that are pretrained using external knowledge on pharmaco-kinetic properties and trial risk from the web. Then the interaction graph module will connect all the embedding via domain knowledge to fully capture various trial components and their complex relations as well as their influences on trial outcomes. Finally, HINT learns a dynamic attentive graph neural network to predict trial outcome. Comprehensive experimental results show that HINT achieves strong predictive performance, obtaining 0.772, 0.607, 0.623, 0.703 on PR-AUC for Phase I, II, III, and indication outcome prediction, respectively. It also consistently outperforms the best baseline method by up to 12.4\% on PR-AUC.
Clinical trials are essential for drug development but often suffer from expensive, inaccurate and insufficient patient recruitment. The core problem of patient-trial matching is to find qualified patients for a trial, where patient information is stored in electronic health records (EHR) while trial eligibility criteria (EC) are described in text documents available on the web. How to represent longitudinal patient EHR? How to extract complex logical rules from EC? Most existing works rely on manual rule-based extraction, which is time consuming and inflexible for complex inference. To address these challenges, we proposed DeepEnroll, a cross-modal inference learning model to jointly encode enrollment criteria (text) and patients records (tabular data) into a shared latent space for matching inference. DeepEnroll applies a pre-trained Bidirectional Encoder Representations from Transformers(BERT) model to encode clinical trial information into sentence embedding. And uses a hierarchical embedding model to represent patient longitudinal EHR. In addition, DeepEnroll is augmented by a numerical information embedding and entailment module to reason over numerical information in both EC and EHR. These encoders are trained jointly to optimize patient-trial matching score. We evaluated DeepEnroll on the trial-patient matching task with demonstrated on real world datasets. DeepEnroll outperformed the best baseline by up to 12.4% in average F1.
The attention mechanism has demonstrated superior performance for inference
over nodes in graph neural networks (GNNs), however, they result in a high
computational burden during both training and inference. We propose FastGAT, a
method to make attention based GNNs lightweight by using spectral
sparsification to generate an optimal pruning of the input graph. This results
in a per-epoch time that is almost linear in the number of graph nodes as
opposed to quadratic. We theoretically prove that spectral sparsification
preserves the features computed by the GAT model, thereby justifying our
algorithm. We experimentally evaluate FastGAT on several large real world graph
datasets for node classification tasks under both inductive and transductive
settings. FastGAT can dramatically reduce (up to \textbf{10x}) the
computational time and memory requirements, allowing the usage of attention
based GNNs on large graphs.
18 Jun 2025
We develop and implement a version of the popular "policytree" method (Athey
and Wager, 2021) using discrete optimisation techniques. We test the
performance of our algorithm in finite samples and find an improvement in the
runtime of optimal policy tree learning by a factor of nearly 50 compared to
the original version. We provide an R package, "fastpolicytree", for public
use.
Patients with low health literacy usually have difficulty understanding medical jargon and the complex structure of professional medical language. Although some studies are proposed to automatically translate expert language into layperson-understandable language, only a few of them focus on both accuracy and readability aspects simultaneously in the clinical domain. Thus, simplification of the clinical language is still a challenging task, but unfortunately, it is not yet fully addressed in previous work. To benchmark this task, we construct a new dataset named MedLane to support the development and evaluation of automated clinical language simplification approaches. Besides, we propose a new model called DECLARE that follows the human annotation procedure and achieves state-of-the-art performance compared with eight strong baselines. To fairly evaluate the performance, we also propose three specific evaluation metrics. Experimental results demonstrate the utility of the annotated MedLane dataset and the effectiveness of the proposed model DECLARE.
 UC Berkeley
UC Berkeley University of VirginiaUniversidad de ChilePontificia Universidad Católica de ChileMontana State UniversityUniversidad de AntofagastaUniversidad Católica del NorteUniversidad de La SerenaIQVIAUniversidad de ConcepciٞnNational Optical-Infrared Astronomy Research Laboratory (NOIRLab)Instituto Multidisciplinario de Investigación y Postgrado
University of VirginiaUniversidad de ChilePontificia Universidad Católica de ChileMontana State UniversityUniversidad de AntofagastaUniversidad Católica del NorteUniversidad de La SerenaIQVIAUniversidad de ConcepciٞnNational Optical-Infrared Astronomy Research Laboratory (NOIRLab)Instituto Multidisciplinario de Investigación y PostgradoWe present the first detailed chemical analysis from APOGEE-2S observations of stars in six regions of recently discovered substructures in the outskirts of the Magellanic Clouds extending to 20 degrees from the LMC center. We also present, for the first time, the metallicity and alpha-abundance radial gradients of the LMC and SMC out to 11 degrees and 6 degrees, respectively. Our chemical tagging includes 13 species including light, alpha, and Fe-peak elements. We find that the abundances of all of these chemical elements in stars populating two regions in the northern periphery - along the northern "stream"-like feature - show good agreement with the chemical patterns of the LMC, and thus likely have an LMC origin. For substructures located in the southern periphery of the LMC, we find more complex chemical and kinematical signatures, indicative of a mix of LMC-like and SMC-like populations. However, the southern region closest to the LMC shows better agreement with the LMC, whereas that closest to the SMC shows a much better agreement with the SMC chemical pattern. When combining this information with 3-D kinematical information for these stars, we conclude that the southern region closest to the LMC has likely an LMC origin, whereas that closest to the SMC has an SMC origin, and the other two southern regions have a mix of LMC and SMC origins. Our results add to the evidence that the southern substructures of the LMC periphery are the product of close interactions between the LMC and SMC, and thus likely hold important clues that can constrain models of their detailed dynamical histories.
Thanks to the increasing availability of drug-drug interactions (DDI)
datasets and large biomedical knowledge graphs (KGs), accurate detection of
adverse DDI using machine learning models becomes possible. However, it remains
largely an open problem how to effectively utilize large and noisy biomedical
KG for DDI detection. Due to its sheer size and amount of noise in KGs, it is
often less beneficial to directly integrate KGs with other smaller but higher
quality data (e.g., experimental data). Most of the existing approaches ignore
KGs altogether. Some try to directly integrate KGs with other data via graph
neural networks with limited success. Furthermore, most previous works focus on
binary DDI prediction whereas the multi-typed DDI pharmacological effect
prediction is a more meaningful but harder task. To fill the gaps, we propose a
new method SumGNN: knowledge summarization graph neural network, which is
enabled by a subgraph extraction module that can efficiently anchor on relevant
subgraphs from a KG, a self-attention based subgraph summarization scheme to
generate a reasoning path within the subgraph, and a multi-channel knowledge
and data integration module that utilizes massive external biomedical knowledge
for significantly improved multi-typed DDI predictions. SumGNN outperforms the
best baseline by up to 5.54\%, and the performance gain is particularly
significant in low data relation types. In addition, SumGNN provides
interpretable prediction via the generated reasoning paths for each prediction.
Massive electronic health records (EHRs) enable the success of learning
accurate patient representations to support various predictive health
applications. In contrast, doctor representation was not well studied despite
that doctors play pivotal roles in healthcare. How to construct the right
doctor representations? How to use doctor representation to solve important
health analytic problems? In this work, we study the problem on {\it clinical
trial recruitment}, which is about identifying the right doctors to help
conduct the trials based on the trial description and patient EHR data of those
doctors. We propose doctor2vec which simultaneously learns 1) doctor
representations from EHR data and 2) trial representations from the description
and categorical information about the trials. In particular, doctor2vec
utilizes a dynamic memory network where the doctor's experience with patients
are stored in the memory bank and the network will dynamically assign weights
based on the trial representation via an attention mechanism. Validated on
large real-world trials and EHR data including 2,609 trials, 25K doctors and
430K patients, doctor2vec demonstrated improved performance over the best
baseline by up to 8.7% in PR-AUC. We also demonstrated that the doctor2vec
embedding can be transferred to benefit data insufficiency settings including
trial recruitment in less populated/newly explored country with 13.7%
improvement or for rare diseases with 8.1% improvement in PR-AUC.
Outlier detection (OD) is a key machine learning (ML) task for identifying abnormal objects from general samples with numerous high-stake applications including fraud detection and intrusion detection. Due to the lack of ground truth labels, practitioners often have to build a large number of unsupervised, heterogeneous models (i.e., different algorithms with varying hyperparameters) for further combination and analysis, rather than relying on a single model. How to accelerate the training and scoring on new-coming samples by outlyingness (referred as prediction throughout the paper) with a large number of unsupervised, heterogeneous OD models? In this study, we propose a modular acceleration system, called SUOD, to address it. The proposed system focuses on three complementary acceleration aspects (data reduction for high-dimensional data, approximation for costly models, and taskload imbalance optimization for distributed environment), while maintaining performance accuracy. Extensive experiments on more than 20 benchmark datasets demonstrate SUOD's effectiveness in heterogeneous OD acceleration, along with a real-world deployment case on fraudulent claim analysis at IQVIA, a leading healthcare firm. We open-source SUOD for reproducibility and accessibility.
With recent advances in natural language processing, rationalization becomes an essential self-explaining diagram to disentangle the black box by selecting a subset of input texts to account for the major variation in prediction. Yet, existing association-based approaches on rationalization cannot identify true rationales when two or more snippets are highly inter-correlated and thus provide a similar contribution to prediction accuracy, so-called spuriousness. To address this limitation, we novelly leverage two causal desiderata, non-spuriousness and efficiency, into rationalization from the causal inference perspective. We formally define a series of probabilities of causation based on a newly proposed structural causal model of rationalization, with its theoretical identification established as the main component of learning necessary and sufficient rationales. The superior performance of the proposed causal rationalization is demonstrated on real-world review and medical datasets with extensive experiments compared to state-of-the-art methods.
We present the first detailed chemical analysis from APOGEE-2S observations of stars in six regions of recently discovered substructures in the outskirts of the Magellanic Clouds extending to 20 degrees from the LMC center. We also present, for the first time, the metallicity and alpha-abundance radial gradients of the LMC and SMC out to 11 degrees and 6 degrees, respectively. Our chemical tagging includes 13 species including light, alpha, and Fe-peak elements. We find that the abundances of all of these chemical elements in stars populating two regions in the northern periphery - along the northern "stream"-like feature - show good agreement with the chemical patterns of the LMC, and thus likely have an LMC origin. For substructures located in the southern periphery of the LMC, we find more complex chemical and kinematical signatures, indicative of a mix of LMC-like and SMC-like populations. However, the southern region closest to the LMC shows better agreement with the LMC, whereas that closest to the SMC shows a much better agreement with the SMC chemical pattern. When combining this information with 3-D kinematical information for these stars, we conclude that the southern region closest to the LMC has likely an LMC origin, whereas that closest to the SMC has an SMC origin, and the other two southern regions have a mix of LMC and SMC origins. Our results add to the evidence that the southern substructures of the LMC periphery are the product of close interactions between the LMC and SMC, and thus likely hold important clues that can constrain models of their detailed dynamical histories.
Background:The electrocardiogram (ECG) is one of the most commonly used
diagnostic tools in medicine and healthcare. Deep learning methods have
achieved promising results on predictive healthcare tasks using ECG signals.
Objective:This paper presents a systematic review of deep learning methods for
ECG data from both modeling and application perspectives. Methods:We extracted
papers that applied deep learning (deep neural network) models to ECG data that
were published between Jan. 1st of 2010 and Feb. 29th of 2020 from Google
Scholar, PubMed, and the DBLP. We then analyzed each article according to three
factors: tasks, models, and data. Finally, we discuss open challenges and
unsolved problems in this area. Results: The total number of papers extracted
was 191. Among these papers, 108 were published after 2019. Different deep
learning architectures have been used in various ECG analytics tasks, such as
disease detection/classification, annotation/localization, sleep staging,
biometric human identification, and denoising. Conclusion: The number of works
on deep learning for ECG data has grown explosively in recent years. Such works
have achieved accuracy comparable to that of traditional feature-based
approaches and ensembles of multiple approaches can achieve even better
results. Specifically, we found that a hybrid architecture of a convolutional
neural network and recurrent neural network ensemble using expert features
yields the best results. However, there are some new challenges and problems
related to interpretability, scalability, and efficiency that must be
addressed. Furthermore, it is also worth investigating new applications from
the perspectives of datasets and methods. Significance: This paper summarizes
existing deep learning research using ECG data from multiple perspectives and
highlights existing challenges and problems to identify potential future
research directions.
Veteran mental health is a significant national problem as large number of
veterans are returning from the recent war in Iraq and continued military
presence in Afghanistan. While significant existing works have investigated
twitter posts-based Post Traumatic Stress Disorder (PTSD) assessment using
blackbox machine learning techniques, these frameworks cannot be trusted by the
clinicians due to the lack of clinical explainability. To obtain the trust of
clinicians, we explore the big question, can twitter posts provide enough
information to fill up clinical PTSD assessment surveys that have been
traditionally trusted by clinicians? To answer the above question, we propose,
LAXARY (Linguistic Analysis-based Exaplainable Inquiry) model, a novel
Explainable Artificial Intelligent (XAI) model to detect and represent PTSD
assessment of twitter users using a modified Linguistic Inquiry and Word Count
(LIWC) analysis. First, we employ clinically validated survey tools for
collecting clinical PTSD assessment data from real twitter users and develop a
PTSD Linguistic Dictionary using the PTSD assessment survey results. Then, we
use the PTSD Linguistic Dictionary along with machine learning model to fill up
the survey tools towards detecting PTSD status and its intensity of
corresponding twitter users. Our experimental evaluation on 210 clinically
validated veteran twitter users provides promising accuracies of both PTSD
classification and its intensity estimation. We also evaluate our developed
PTSD Linguistic Dictionary's reliability and validity.
There are no more papers matching your filters at the moment.