AstraZeneca
Latent Refinement Decoding (LRD) enhances diffusion-based language models by introducing a two-stage decoding framework that refines belief states through continuous embedding updates and adaptive sampling. This approach consistently improves generation accuracy and achieves inference speedups of up to 10.6x across code generation and mathematical reasoning tasks.
08 Oct 2025
Researchers at Chalmers University of Technology developed Transferable Implicit Transfer Operators (TITO), a deep generative model that bridges femtosecond to nanosecond timescales in molecular dynamics simulations. TITO accurately reproduces both equilibrium and kinetic properties of molecular systems, achieving up to a 15,000-fold acceleration over conventional molecular dynamics while demonstrating transferability to unseen molecules.
We introduce FLOWR, a novel structure-based framework for the generation and
optimization of three-dimensional ligands. FLOWR integrates continuous and
categorical flow matching with equivariant optimal transport, enhanced by an
efficient protein pocket conditioning. Alongside FLOWR, we present SPINDR, a
thoroughly curated dataset comprising ligand-pocket co-crystal complexes
specifically designed to address existing data quality issues. Empirical
evaluations demonstrate that FLOWR surpasses current state-of-the-art
diffusion- and flow-based methods in terms of PoseBusters-validity, pose
accuracy, and interaction recovery, while offering a significant inference
speedup, achieving up to 70-fold faster performance. In addition, we introduce
FLOWR:multi, a highly accurate multi-purpose model allowing for the targeted
sampling of novel ligands that adhere to predefined interaction profiles and
chemical substructures for fragment-based design without the need of
re-training or any re-sampling strategies
Methods for jointly generating molecular graphs along with their 3D
conformations have gained prominence recently due to their potential impact on
structure-based drug design. Current approaches, however, often suffer from
very slow sampling times or generate molecules with poor chemical validity.
Addressing these limitations, we propose Semla, a scalable E(3)-equivariant
message passing architecture. We further introduce an unconditional 3D
molecular generation model, SemlaFlow, which is trained using equivariant flow
matching to generate a joint distribution over atom types, coordinates, bond
types and formal charges. Our model produces state-of-the-art results on
benchmark datasets with as few as 20 sampling steps, corresponding to a two
order-of-magnitude speedup compared to state-of-the-art. Furthermore, we
highlight limitations of current evaluation methods for 3D generation and
propose new benchmark metrics for unconditional molecular generators. Finally,
using these new metrics, we compare our model's ability to generate high
quality samples against current approaches and further demonstrate SemlaFlow's
strong performance.
Researchers introduce Compositional Generative Flows (CGFlow), a framework that integrates compositional generation with continuous feature modeling, and apply it as 3DSynthFlow for target-based drug design. This model co-designs 3D molecular structures and their synthesis pathways, achieving state-of-the-art binding affinity (e.g., -11.96 kcal/mol on ADRB2) and improved sampling efficiency (4.2x over RxnFlow) while maintaining high synthesizability (68.58% success rate).
MADRIGAL, a multimodal AI model developed by researchers including those from Harvard Medical School and AstraZeneca, predicts clinical outcomes of drug combinations using diverse preclinical data while robustly handling missing information. The model significantly improves prediction of adverse events and patient-specific treatment responses, demonstrating strong alignment with real-world clinical trial data and electronic health records.
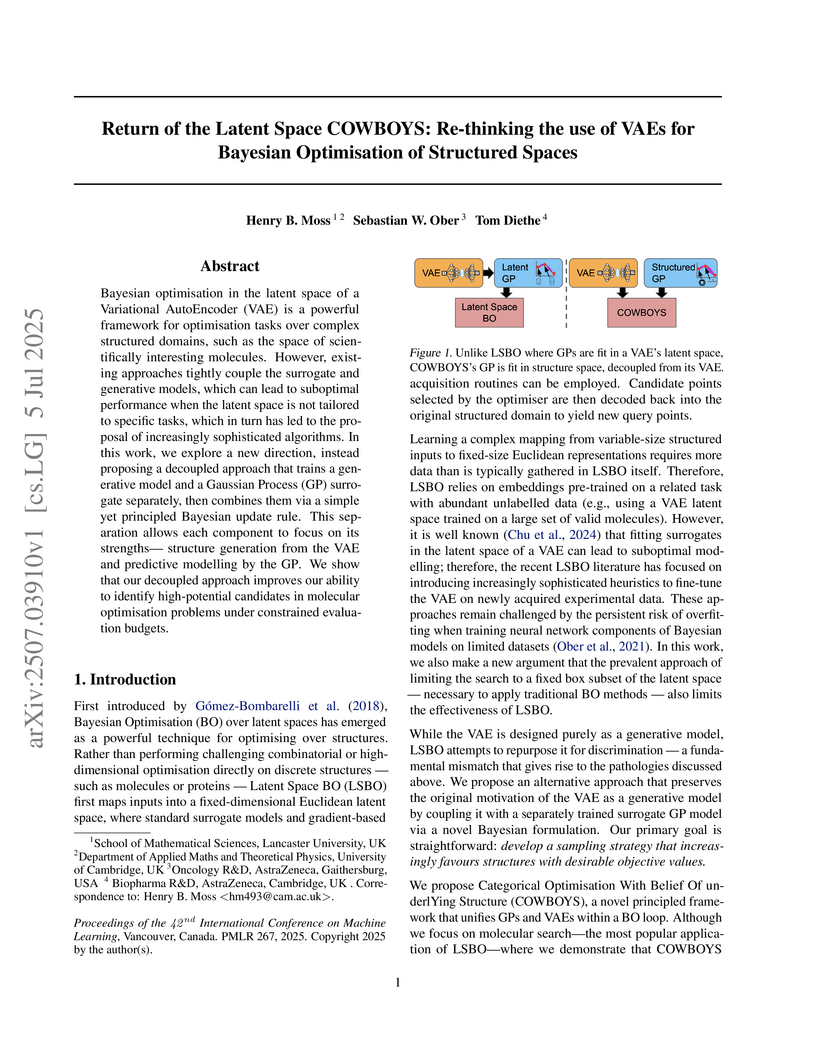
COWBOYS (Categorical Optimisation With Belief Of underlYing Structure), developed by researchers including those from AstraZeneca, introduces a Bayesian Optimization framework that re-thinks the use of Variational AutoEncoders (VAEs) by decoupling them from Gaussian Process (GP) surrogate modeling, allowing GPs to operate directly in the original structured data space. This approach achieves marked improvements in sample efficiency, particularly in low-data regimes, across various molecular design benchmarks compared to existing Latent Space Bayesian Optimization methods.
Over the last few years, the Shapley value, a solution concept from cooperative game theory, has found numerous applications in machine learning. In this paper, we first discuss fundamental concepts of cooperative game theory and axiomatic properties of the Shapley value. Then we give an overview of the most important applications of the Shapley value in machine learning: feature selection, explainability, multi-agent reinforcement learning, ensemble pruning, and data valuation. We examine the most crucial limitations of the Shapley value and point out directions for future research.
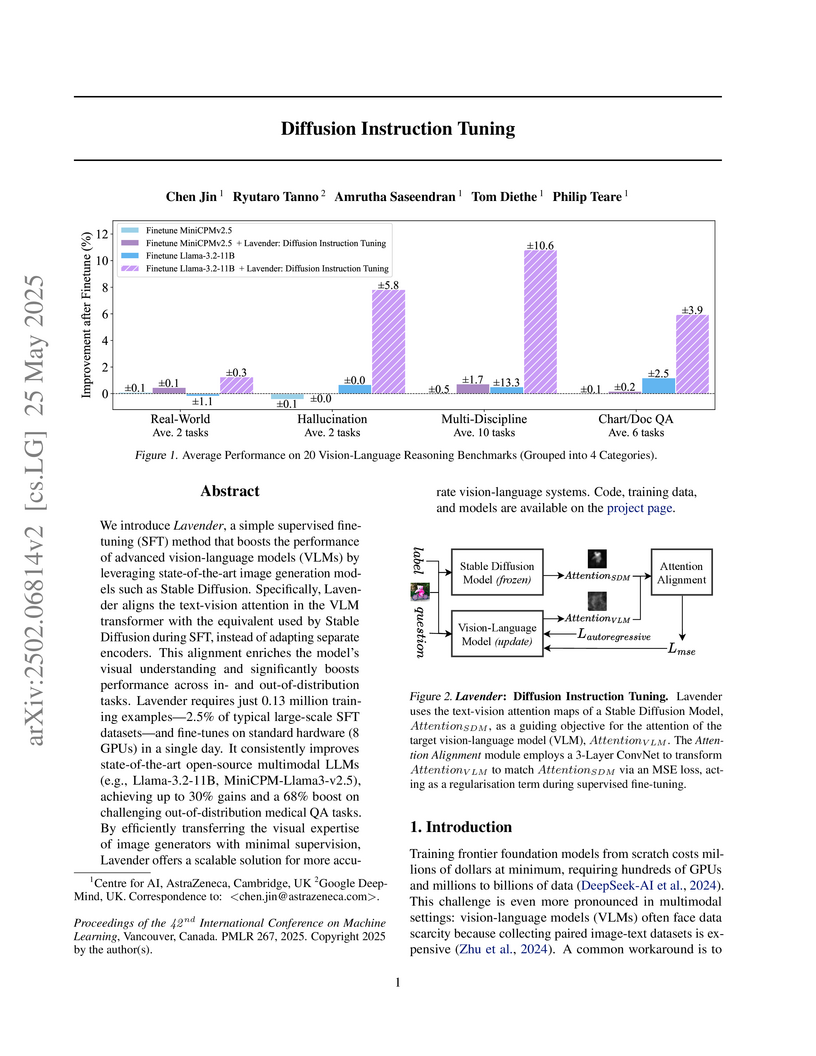
We introduce Lavender, a simple supervised fine-tuning (SFT) method that
boosts the performance of advanced vision-language models (VLMs) by leveraging
state-of-the-art image generation models such as Stable Diffusion.
Specifically, Lavender aligns the text-vision attention in the VLM transformer
with the equivalent used by Stable Diffusion during SFT, instead of adapting
separate encoders. This alignment enriches the model's visual understanding and
significantly boosts performance across in- and out-of-distribution tasks.
Lavender requires just 0.13 million training examples, 2.5% of typical
large-scale SFT datasets, and fine-tunes on standard hardware (8 GPUs) in a
single day. It consistently improves state-of-the-art open-source multimodal
LLMs (e.g., Llama-3.2-11B, MiniCPM-Llama3-v2.5), achieving up to 30% gains and
a 68% boost on challenging out-of-distribution medical QA tasks. By efficiently
transferring the visual expertise of image generators with minimal supervision,
Lavender offers a scalable solution for more accurate vision-language systems.
All code, training data, and models will be shared at
this https URL
Researchers at the University of Cambridge and AstraZeneca empirically investigated adaptive readout functions for Graph Neural Networks (GNNs). Their study demonstrated that these differentiable readouts consistently improved performance across over 40 datasets, often converging faster, and achieved substantial gains (e.g., R2 from 0.33 to 0.78) on multi-million scale molecular datasets in drug discovery.
21 Aug 2025
Large-language models (LLMs) and agentic systems present exciting opportunities to accelerate drug discovery. In this study, we examine the modularity of LLM-based agentic systems for drug discovery, i.e., whether parts of the system such as the LLM and type of agent are interchangeable, a topic that has received limited attention in drug discovery. We compare the performance of different LLMs and the effectiveness of tool-calling agents versus code-generating agents. Our case study, comparing performance in orchestrating tools for chemistry and drug discovery using an LLM-as-a-judge score, shows that Claude-3.5-Sonnet, Claude-3.7-Sonnet and GPT-4o outperform alternative language models such as Llama-3.1-8B, Llama-3.1-70B, GPT-3.5-Turbo, and Nova-Micro. Although we confirm that code-generating agents outperform the tool-calling ones on average, we show that this is highly question- and model-dependent. Furthermore, the impact of replacing system prompts is dependent on the question and model, underscoring that even in this particular domain one cannot just replace components of the system without re-engineering. Our study highlights the necessity of further research into the modularity of agentic systems to enable the development of reliable and modular solutions for real-world problems.
Smart training set selections procedures enable the reduction of data needs and improves predictive robustness in machine learning problems relevant to chemistry. We introduce Gradient Guided Furthest Point Sampling (GGFPS), a simple extension of Furthest Point Sampling (FPS) that leverages molecular force norms to guide efficient sampling of configurational spaces of molecules. Numerical evidence is presented for a toy-system (Styblinski-Tang function) as well as for molecular dynamics trajectories from the MD17 dataset. Compared to FPS and uniform sampling, our numerical results indicate superior data efficiency and robustness when using GGFPS. Distribution analysis of the MD17 data suggests that FPS systematically under-samples equilibrium geometries, resulting in large test errors for relaxed structures. GGFPS cures this artifact and (i) enables up to two fold reductions in training cost without sacrificing predictive accuracy compared to FPS in the 2-dimensional Styblinksi-Tang system, (ii) systematically lowers prediction errors for equilibrium as well as strained structures in MD17, and (iii) systematically decreases prediction error variances across all of the MD17 configuration spaces. These results suggest that gradient-aware sampling methods hold great promise as effective training set selection tools, and that naive use of FPS may result in imbalanced training and inconsistent prediction outcomes.
Segment Anyword introduces a training-free framework for open-set language-grounded image segmentation, leveraging pre-trained diffusion models and linguistic knowledge. This approach achieves strong performance on benchmarks like Pascal Context 59 (52.5 mIoU) and gRefCOCO (67.73 cIoU) without requiring model training or fine-tuning.
Large Language Models (LLMs) often hallucinate, producing unfaithful or factually incorrect outputs by misrepresenting the provided context or incorrectly recalling internal knowledge. Recent studies have identified specific attention heads within the Transformer architecture, known as retrieval heads, responsible for extracting relevant contextual information. We hypothesise that masking these retrieval heads can induce hallucinations and that contrasting the outputs of the base LLM and the masked LLM can reduce hallucinations. To this end, we propose Decoding by Contrasting Retrieval Heads (DeCoRe), a novel training-free decoding strategy that amplifies information found in the context and model parameters. DeCoRe mitigates potentially hallucinated responses by dynamically contrasting the outputs of the base LLM and the masked LLM, using conditional entropy as a guide. Our extensive experiments confirm that DeCoRe significantly improves performance on tasks requiring high contextual faithfulness, such as summarisation (XSum by 18.6%), instruction following (MemoTrap by 10.9%), and open-book question answering (NQ-Open by 2.4% and NQ-Swap by 5.5%).
14 Aug 2025
Estimating treatment effects (TE) from observational data is a critical yet complex task in many fields, from healthcare and economics to public policy. While recent advances in machine learning and causal inference have produced powerful estimation techniques, their adoption remains limited due to the need for deep expertise in causal assumptions, adjustment strategies, and model selection. In this paper, we introduce CATE-B, an open-source co-pilot system that uses large language models (LLMs) within an agentic framework to guide users through the end-to-end process of treatment effect estimation. CATE-B assists in (i) constructing a structural causal model via causal discovery and LLM-based edge orientation, (ii) identifying robust adjustment sets through a novel Minimal Uncertainty Adjustment Set criterion, and (iii) selecting appropriate regression methods tailored to the causal structure and dataset characteristics. To encourage reproducibility and evaluation, we release a suite of benchmark tasks spanning diverse domains and causal complexities. By combining causal inference with intelligent, interactive assistance, CATE-B lowers the barrier to rigorous causal analysis and lays the foundation for a new class of benchmarks in automated treatment effect estimation.
11 May 2025
The first WARA Robotics Mobile Manipulation Challenge, held in December 2024
at ABB Corporate Research in V\"aster{\aa}s, Sweden, addressed the automation
of task-intensive and repetitive manual labor in laboratory environments -
specifically the transport and cleaning of glassware. Designed in collaboration
with AstraZeneca, the challenge invited academic teams to develop autonomous
robotic systems capable of navigating human-populated lab spaces and performing
complex manipulation tasks, such as loading items into industrial dishwashers.
This paper presents an overview of the challenge setup, its industrial
motivation, and the four distinct approaches proposed by the participating
teams. We summarize lessons learned from this edition and propose improvements
in design to enable a more effective second iteration to take place in 2025.
The initiative bridges an important gap in effective academia-industry
collaboration within the domain of autonomous mobile manipulation systems by
promoting the development and deployment of applied robotic solutions in
real-world laboratory contexts.
SubTab, developed by researchers at AstraZeneca, introduces a self-supervised learning framework for tabular data that subsets features to create multiple views, enabling an autoencoder to reconstruct the entire input from partial information. This method achieved 98.31% accuracy on tabular MNIST and consistently outperformed other self-supervised baselines on diverse real-world datasets like TCGA and Obesity data.
FlexiFlow introduces a generative model that simultaneously produces molecular graphs and their low-energy conformational ensembles, achieving state-of-the-art molecular generation metrics and high-quality, diverse conformers efficiently, even for protein-conditioned ligand design.
The ASKCOS project, developed at MIT with industrial collaboration, is an open-source software suite providing a comprehensive platform for data-driven chemical synthesis planning. It integrates various machine learning models and search algorithms for multi-step retrosynthesis, reaction condition recommendation, and outcome prediction, assisting chemists in daily tasks and accelerating chemical discovery.
A fundamental challenge in protein design is the trade-off between generating structural diversity while preserving motif biological function. Current state-of-the-art methods, such as partial diffusion in RFdiffusion, often fail to resolve this trade-off: small perturbations yield motifs nearly identical to the native structure, whereas larger perturbations violate the geometric constraints necessary for biological function. We introduce Protein Generation with Embedding Learning (PGEL), a general framework that learns high-dimensional embeddings encoding sequence and structural features of a target motif in the representation space of a diffusion model's frozen denoiser, and then enhances motif diversity by introducing controlled perturbations in the embedding space. PGEL is thus able to loosen geometric constraints while satisfying typical design metrics, leading to more diverse yet viable structures. We demonstrate PGEL on three representative cases: a monomer, a protein-protein interface, and a cancer-related transcription factor complex. In all cases, PGEL achieves greater structural diversity, better designability, and improved self-consistency, as compared to partial diffusion. Our results establish PGEL as a general strategy for embedding-driven protein generation allowing for systematic, viable diversification of functional motifs.
There are no more papers matching your filters at the moment.