Federal University of Minas Gerais
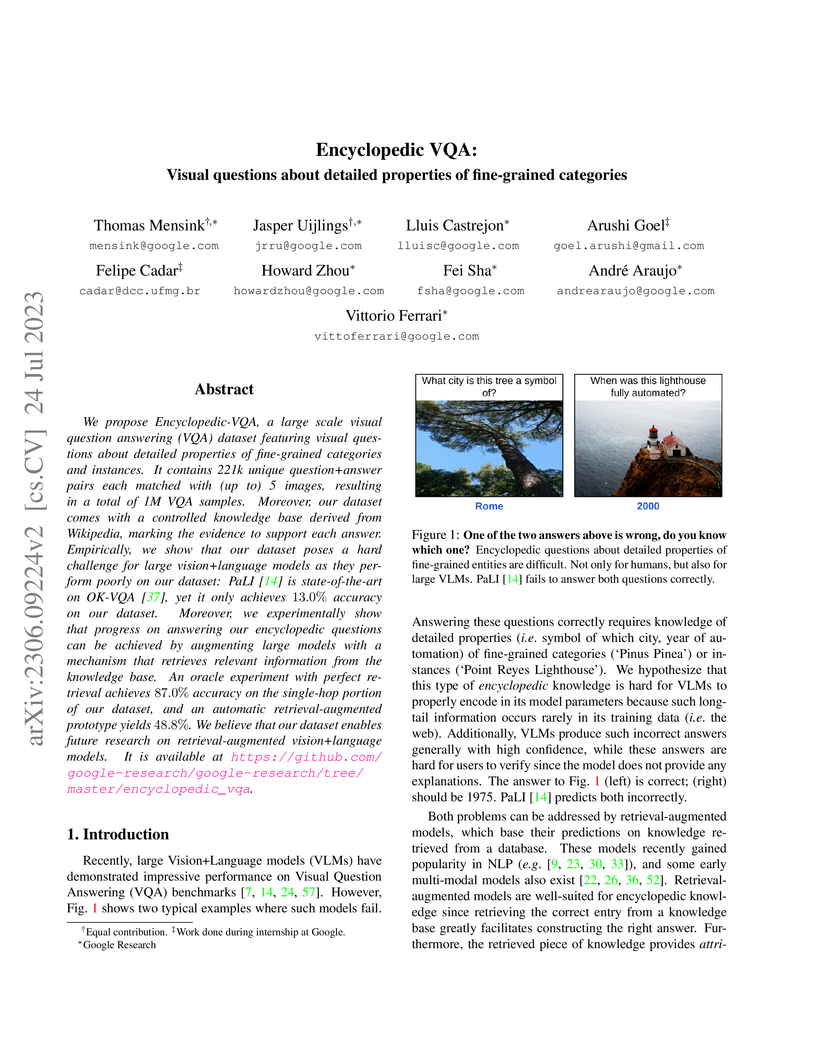
Encyclopedic VQA introduces a 1 million-sample dataset with visual questions requiring detailed properties of fine-grained categories, revealing that state-of-the-art Vision-Language Models struggle with such knowledge unless augmented with external, attributable retrieval from a controlled knowledge base. This work highlights the necessity of knowledge retrieval for verifiable and trustworthy AI systems in knowledge-intensive VQA.
With the rapid growth in the number of Large Language Models (LLMs), there
has been a recent interest in LLM routing, or directing queries to the cheapest
LLM that can deliver a suitable response. We conduct a minimax analysis of the
routing problem, providing a lower bound and finding that a simple router that
predicts both cost and accuracy for each question can be minimax optimal.
Inspired by this, we introduce CARROT, a Cost AwaRe Rate Optimal rouTer that
selects a model based on estimates of the models' cost and performance.
Alongside CARROT, we also introduce the Smart Price-aware ROUTing (SPROUT)
dataset to facilitate routing on a wide spectrum of queries with the latest
state-of-the-art LLMs. Using SPROUT and prior benchmarks such as Routerbench
and open-LLM-leaderboard-v2 we empirically validate CARROT's performance
against several alternative routers.
PromptEval, developed by researchers including those from the MIT-IBM Watson AI Lab, introduces an efficient method based on Item Response Theory to estimate the full performance distribution of Large Language Models across a large pool of prompt templates. It provides accurate performance quantiles with a budget equivalent to just two single-prompt evaluations, addressing the high computational cost and prompt sensitivity of current LLM evaluation practices.
Large Language Models (LLMs) have gained attention for addressing coding problems, but their effectiveness in fixing code maintainability remains unclear. This study evaluates LLMs capability to resolve 127 maintainability issues from 10 GitHub repositories. We use zero-shot prompting for Copilot Chat and Llama 3.1, and few-shot prompting with Llama only. The LLM-generated solutions are assessed for compilation errors, test failures, and new maintainability problems. Llama with few-shot prompting successfully fixed 44.9% of the methods, while Copilot Chat and Llama zero-shot fixed 32.29% and 30%, respectively. However, most solutions introduced errors or new maintainability issues. We also conducted a human study with 45 participants to evaluate the readability of 51 LLM-generated solutions. The human study showed that 68.63% of participants observed improved readability. Overall, while LLMs show potential for fixing maintainability issues, their introduction of errors highlights their current limitations.
Recent advancements in super-resolution for License Plate Recognition (LPR)
have sought to address challenges posed by low-resolution (LR) and degraded
images in surveillance, traffic monitoring, and forensic applications. However,
existing studies have relied on private datasets and simplistic degradation
models. To address this gap, we introduce UFPR-SR-Plates, a novel dataset
containing 10,000 tracks with 100,000 paired low and high-resolution license
plate images captured under real-world conditions. We establish a benchmark
using multiple sequential LR and high-resolution (HR) images per vehicle --
five of each -- and two state-of-the-art models for super-resolution of license
plates. We also investigate three fusion strategies to evaluate how combining
predictions from a leading Optical Character Recognition (OCR) model for
multiple super-resolved license plates enhances overall performance. Our
findings demonstrate that super-resolution significantly boosts LPR
performance, with further improvements observed when applying majority
vote-based fusion techniques. Specifically, the Layout-Aware and
Character-Driven Network (LCDNet) model combined with the Majority Vote by
Character Position (MVCP) strategy led to the highest recognition rates,
increasing from 1.7% with low-resolution images to 31.1% with super-resolution,
and up to 44.7% when combining OCR outputs from five super-resolved images.
These findings underscore the critical role of super-resolution and temporal
information in enhancing LPR accuracy under real-world, adverse conditions. The
proposed dataset is publicly available to support further research and can be
accessed at: this https URL
Despite significant advancements in License Plate Recognition (LPR) through deep learning, most improvements rely on high-resolution images with clear characters. This scenario does not reflect real-world conditions where traffic surveillance often captures low-resolution and blurry images. Under these conditions, characters tend to blend with the background or neighboring characters, making accurate LPR challenging. To address this issue, we introduce a novel loss function, Layout and Character Oriented Focal Loss (LCOFL), which considers factors such as resolution, texture, and structural details, as well as the performance of the LPR task itself. We enhance character feature learning using deformable convolutions and shared weights in an attention module and employ a GAN-based training approach with an Optical Character Recognition (OCR) model as the discriminator to guide the super-resolution process. Our experimental results show significant improvements in character reconstruction quality, outperforming two state-of-the-art methods in both quantitative and qualitative measures. Our code is publicly available at this https URL
Researchers developed the DivShift framework and DivShift-NAWC dataset to quantitatively assess how ecologically and sociopolitically relevant biases in large-scale volunteer biodiversity data affect species recognition model performance. The study revealed that models generalize better than expected from label shifts, but significant performance disparities persist across various bias types like spatial and observer differences.
Software development increasingly depends on libraries and frameworks to
increase productivity and reduce time-to-market. Despite this fact, we still
lack techniques to assess developers expertise in widely popular libraries and
frameworks. In this paper, we evaluate the performance of unsupervised (based
on clustering) and supervised machine learning classifiers (Random Forest and
SVM) to identify experts in three popular JavaScript libraries: facebook/react,
mongodb/node-mongodb, and socketio/socket.io. First, we collect 13 features
about developers activity on GitHub projects, including commits on source code
files that depend on these libraries. We also build a ground truth including
the expertise of 575 developers on the studied libraries, as self-reported by
them in a survey. Based on our findings, we document the challenges of using
machine learning classifiers to predict expertise in software libraries, using
features extracted from GitHub. Then, we propose a method to identify library
experts based on clustering feature data from GitHub; by triangulating the
results of this method with information available on Linkedin profiles, we show
that it is able to recommend dozens of GitHub users with evidences of being
experts in the studied JavaScript libraries. We also provide a public dataset
with the expertise of 575 developers on the studied libraries.
25 Sep 2024
Despite being introduced only a few years ago, Large Language Models (LLMs) are already widely used by developers for code generation. However, their application in automating other Software Engineering activities remains largely unexplored. Thus, in this paper, we report the first results of a study in which we are exploring the use of ChatGPT to support API migration tasks, an important problem that demands manual effort and attention from developers. Specifically, in the paper, we share our initial results involving the use of ChatGPT to migrate a client application to use a newer version of SQLAlchemy, an ORM (Object Relational Mapping) library widely used in Python. We evaluate the use of three types of prompts (Zero-Shot, One-Shot, and Chain Of Thoughts) and show that the best results are achieved by the One-Shot prompt, followed by the Chain Of Thoughts. Particularly, with the One-Shot prompt we were able to successfully migrate all columns of our target application and upgrade its code to use new functionalities enabled by SQLAlchemy's latest version, such as Python's asyncio and typing modules, while preserving the original code behavior.
This paper introduces a novel Graph Neural Network (GNN) architecture for time series classification, based on visibility graph representations. Traditional time series classification methods often struggle with high computational complexity and inadequate capture of spatio-temporal dynamics. By representing time series as visibility graphs, it is possible to encode both spatial and temporal dependencies inherent to time series data, while being computationally efficient. Our architecture is fully modular, enabling flexible experimentation with different models and representations. We employ directed visibility graphs encoded with in-degree and PageRank features to improve the representation of time series, ensuring efficient computation while enhancing the model's ability to capture long-range dependencies in the data. We show the robustness and generalization capability of the proposed architecture across a diverse set of classification tasks and against a traditional model. Our work represents a significant advancement in the application of GNNs for time series analysis, offering a powerful and flexible framework for future research and practical implementations.
22 Sep 2024
Robots must operate safely when deployed in novel and human-centered environments, like homes. Current safe control approaches typically assume that the safety constraints are known a priori, and thus, the robot can pre-compute a corresponding safety controller. While this may make sense for some safety constraints (e.g., avoiding collision with walls by analyzing a floor plan), other constraints are more complex (e.g., spills), inherently personal, context-dependent, and can only be identified at deployment time when the robot is interacting in a specific environment and with a specific person (e.g., fragile objects, expensive rugs). Here, language provides a flexible mechanism to communicate these evolving safety constraints to the robot. In this work, we use vision language models (VLMs) to interpret language feedback and the robot's image observations to continuously update the robot's representation of safety constraints. With these inferred constraints, we update a Hamilton-Jacobi reachability safety controller online via efficient warm-starting techniques. Through simulation and hardware experiments, we demonstrate the robot's ability to infer and respect language-based safety constraints with the proposed approach.
Automated news credibility and fact-checking at scale require accurately predicting news factuality and media bias. This paper introduces a large sentence-level dataset, titled "FactNews", composed of 6,191 sentences expertly annotated according to factuality and media bias definitions proposed by AllSides. We use FactNews to assess the overall reliability of news sources, by formulating two text classification problems for predicting sentence-level factuality of news reporting and bias of media outlets. Our experiments demonstrate that biased sentences present a higher number of words compared to factual sentences, besides having a predominance of emotions. Hence, the fine-grained analysis of subjectivity and impartiality of news articles provided promising results for predicting the reliability of media outlets. Finally, due to the severity of fake news and political polarization in Brazil, and the lack of research for Portuguese, both dataset and baseline were proposed for Brazilian Portuguese.
21 Mar 2021
Code review is a key development practice that contributes to improve software quality and to foster knowledge sharing among developers. However, code review usually takes time and demands detailed and time-consuming analysis of textual diffs. Particularly, detecting refactorings during code reviews is not a trivial task, since they are not explicitly represented in diffs. For example, a Move Function refactoring is represented by deleted (-) and added lines (+) of code which can be located in different and distant source code files. To tackle this problem, we introduce RAID, a refactoring-aware and intelligent diff tool. Besides proposing an architecture for RAID, we implemented a Chrome browser plug-in that supports our solution. Then, we conducted a field experiment with eight professional developers who used RAID for three months. We concluded that RAID can reduce the cognitive effort required for detecting and reviewing refactorings in textual diff. Besides documenting refactorings in diffs, RAID reduces the number of lines required for reviewing such operations. For example, the median number of lines to be reviewed decreases from 14.5 to 2 lines in the case of move refactorings and from 113 to 55 lines in the case of extractions.
In the last few years thousands of scientific papers have investigated sentiment analysis, several startups that measure opinions on real data have emerged and a number of innovative products related to this theme have been developed. There are multiple methods for measuring sentiments, including lexical-based and supervised machine learning methods. Despite the vast interest on the theme and wide popularity of some methods, it is unclear which one is better for identifying the polarity (i.e., positive or negative) of a message. Accordingly, there is a strong need to conduct a thorough apple-to-apple comparison of sentiment analysis methods, \textit{as they are used in practice}, across multiple datasets originated from different data sources. Such a comparison is key for understanding the potential limitations, advantages, and disadvantages of popular methods. This article aims at filling this gap by presenting a benchmark comparison of twenty-four popular sentiment analysis methods (which we call the state-of-the-practice methods). Our evaluation is based on a benchmark of eighteen labeled datasets, covering messages posted on social networks, movie and product reviews, as well as opinions and comments in news articles. Our results highlight the extent to which the prediction performance of these methods varies considerably across datasets. Aiming at boosting the development of this research area, we open the methods' codes and datasets used in this article, deploying them in a benchmark system, which provides an open API for accessing and comparing sentence-level sentiment analysis methods.
Neoadjuvant chemotherapy (NAC) response prediction for triple negative breast
cancer (TNBC) patients is a challenging task clinically as it requires
understanding complex histology interactions within the tumor microenvironment
(TME). Digital whole slide images (WSIs) capture detailed tissue information,
but their giga-pixel size necessitates computational methods based on multiple
instance learning, which typically analyze small, isolated image tiles without
the spatial context of the TME. To address this limitation and incorporate TME
spatial histology interactions in predicting NAC response for TNBC patients, we
developed a histology context-aware transformer graph convolution network
(NACNet). Our deep learning method identifies the histopathological labels on
individual image tiles from WSIs, constructs a spatial TME graph, and
represents each node with features derived from tissue texture and social
network analysis. It predicts NAC response using a transformer graph
convolution network model enhanced with graph isomorphism network layers. We
evaluate our method with WSIs of a cohort of TNBC patient (N=105) and compared
its performance with multiple state-of-the-art machine learning and deep
learning models, including both graph and non-graph approaches. Our NACNet
achieves 90.0% accuracy, 96.0% sensitivity, 88.0% specificity, and an AUC of
0.82, through eight-fold cross-validation, outperforming baseline models. These
comprehensive experimental results suggest that NACNet holds strong potential
for stratifying TNBC patients by NAC response, thereby helping to prevent
overtreatment, improve patient quality of life, reduce treatment cost, and
enhance clinical outcomes, marking an important advancement toward personalized
breast cancer treatment.
In this paper, we tackle Automatic Meter Reading (AMR) by leveraging the high
capability of Convolutional Neural Networks (CNNs). We design a two-stage
approach that employs the Fast-YOLO object detector for counter detection and
evaluates three different CNN-based approaches for counter recognition. In the
AMR literature, most datasets are not available to the research community since
the images belong to a service company. In this sense, we introduce a new
public dataset, called UFPR-AMR dataset, with 2,000 fully and manually
annotated images. This dataset is, to the best of our knowledge, three times
larger than the largest public dataset found in the literature and contains a
well-defined evaluation protocol to assist the development and evaluation of
AMR methods. Furthermore, we propose the use of a data augmentation technique
to generate a balanced training set with many more examples to train the CNN
models for counter recognition. In the proposed dataset, impressive results
were obtained and a detailed speed/accuracy trade-off evaluation of each model
was performed. In a public dataset, state-of-the-art results were achieved
using less than 200 images for training.
Next-generation touristic services will rely on the advanced mobile networks'
high bandwidth and low latency and the Multi-access Edge Computing (MEC)
paradigm to provide fully immersive mobile experiences. As an integral part of
travel planning systems, recommendation algorithms devise personalized tour
itineraries for individual users considering the popularity of a city's Points
of Interest (POIs) as well as the tourist preferences and constraints. However,
in the context of next-generation touristic services, recommendation algorithms
should also consider the applications (e.g., social network, mobile video
streaming, mobile augmented reality) the tourist will consume in the POIs and
the quality in which the MEC infrastructure will deliver such applications. In
this paper, we address the joint problem of recommending personalized tour
itineraries for tourists and efficiently allocating MEC resources for advanced
touristic applications. We formulate an optimization problem that maximizes the
itinerary of individual tourists while optimizing the resource allocation at
the network edge. We then propose an exact algorithm that quickly solves the
problem optimally, considering instances of realistic size. Using a real-world
location-based photo-sharing database, we conduct and present an exploratory
analysis to understand preferences and users' visiting patterns. Using this
understanding, we propose a methodology to identify user interest in
applications. Finally, we evaluate our algorithm using this dataset. Results
show that our algorithm outperforms a modified version of a state-of-the-art
solution for personalized tour itinerary recommendation, demonstrating gains up
to 11% for resource allocation efficiency and 40% for user experience. In
addition, our algorithm performs similarly to the modified state-of-the-art
solution regarding traditional itinerary recommendation metrics.
24 Jul 2024
This is the first work to investigate the effectiveness of BERT-based contextual embeddings in active learning (AL) tasks on cold-start scenarios, where traditional fine-tuning is infeasible due to the absence of labeled data. Our primary contribution is the proposal of a more robust fine-tuning pipeline - DoTCAL - that diminishes the reliance on labeled data in AL using two steps: (1) fully leveraging unlabeled data through domain adaptation of the embeddings via masked language modeling and (2) further adjusting model weights using labeled data selected by AL. Our evaluation contrasts BERT-based embeddings with other prevalent text representation paradigms, including Bag of Words (BoW), Latent Semantic Indexing (LSI), and FastText, at two critical stages of the AL process: instance selection and classification. Experiments conducted on eight ATC benchmarks with varying AL budgets (number of labeled instances) and number of instances (about 5,000 to 300,000) demonstrate DoTCAL's superior effectiveness, achieving up to a 33% improvement in Macro-F1 while reducing labeling efforts by half compared to the traditional one-step method. We also found that in several tasks, BoW and LSI (due to information aggregation) produce results superior (up to 59% ) to BERT, especially in low-budget scenarios and hard-to-classify tasks, which is quite surprising.
The accuracy of a classifier, when performing Pattern recognition, is mostly
tied to the quality and representativeness of the input feature vector. Feature
Selection is a process that allows for representing information properly and
may increase the accuracy of a classifier. This process is responsible for
finding the best possible features, thus allowing us to identify to which class
a pattern belongs. Feature selection methods can be categorized as Filters,
Wrappers, and Embed. This paper presents a survey on some Filters and Wrapper
methods for handcrafted feature selection. Some discussions, with regard to the
data structure, processing time, and ability to well represent a feature
vector, are also provided in order to explicitly show how appropriate some
methods are in order to perform feature selection. Therefore, the presented
feature selection methods can be accurate and efficient if applied considering
their positives and negatives, finding which one fits best the problem's domain
may be the hardest task.
01 Aug 2019
This work uses vector field inequalities (VFI) to prevent robot
self-collisions and collisions with the workspace. Differently from previous
approaches, the method is suitable for both velocity and torque-actuated
robots. We propose a new distance function and its corresponding Jacobian in
order to generate a VFI to limit the angle between two Pl\"ucker lines. This
new VFI is used to prevent both undesired end-effector orientations and
violation of joints limits. The proposed method is evaluated in a realistic
simulation and on a real humanoid robot, showing that all constraints are
respected while the robot performs a manipulation task.
There are no more papers matching your filters at the moment.