Weill Cornell Medicine
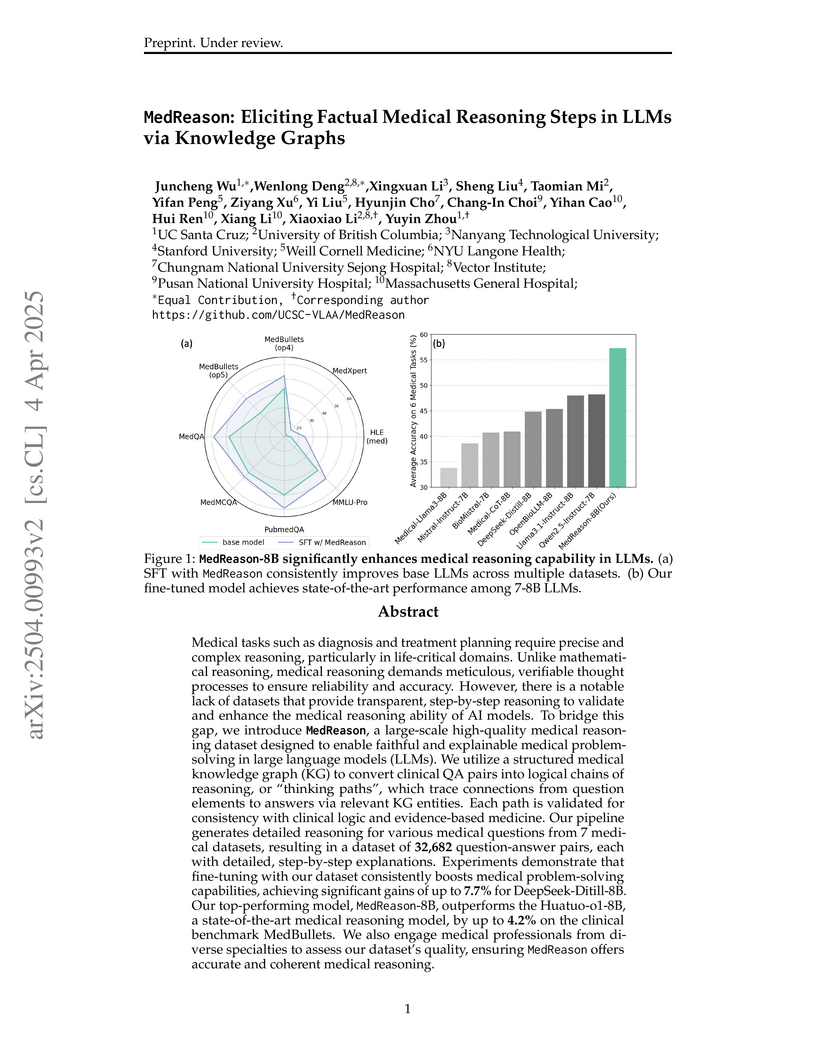
MedReason generates a high-quality, factually grounded Chain-of-Thought (CoT) dataset for Large Language Models (LLMs) by integrating medical knowledge graphs. This approach improves LLM factual accuracy, explainability, and overall performance in medical question-answering, achieving state-of-the-art results for 7-8B parameter models.
Heidelberg University UCLAUniversity of ZurichGeorge Washington University
UCLAUniversity of ZurichGeorge Washington University University of California, Irvine
University of California, Irvine Stanford University
Stanford University University of Michigan
University of Michigan Cornell University
Cornell University University of California, San Diego
University of California, San Diego McGill University
McGill University Northwestern UniversityUniversity of Missouri
Northwestern UniversityUniversity of Missouri University of PennsylvaniaMassachusetts General HospitalUppsala University
University of PennsylvaniaMassachusetts General HospitalUppsala University Duke UniversityHelmholtz MunichMayo ClinicWeill Cornell MedicineIran University of Medical SciencesChildren’s National HospitalUniversity Hospital UlmNational Institute of HealthSage BionetworksUniversity of Rochester Medical CenterMercy Catholic Medical CenterKlinikum rechts der Isar, Technical University of MunichThe Children’s Hospital of PhiladelphiaC.M.H. Lahore Medical College & Institute of DentistryRoss University School of MedicineNassau Medical CenterUniversity of LavalIndiana UniveristyHospital Garcia de Orta - Unidade Local de Sa ́ude de Almada Seixal, EPEScripps Clinic Medical GroupHospital da Luz LisboaVA San Diego Medical CenterASST Ovest Milanese - Legnano
Duke UniversityHelmholtz MunichMayo ClinicWeill Cornell MedicineIran University of Medical SciencesChildren’s National HospitalUniversity Hospital UlmNational Institute of HealthSage BionetworksUniversity of Rochester Medical CenterMercy Catholic Medical CenterKlinikum rechts der Isar, Technical University of MunichThe Children’s Hospital of PhiladelphiaC.M.H. Lahore Medical College & Institute of DentistryRoss University School of MedicineNassau Medical CenterUniversity of LavalIndiana UniveristyHospital Garcia de Orta - Unidade Local de Sa ́ude de Almada Seixal, EPEScripps Clinic Medical GroupHospital da Luz LisboaVA San Diego Medical CenterASST Ovest Milanese - Legnano
 UCLAUniversity of ZurichGeorge Washington University
UCLAUniversity of ZurichGeorge Washington University University of California, Irvine
University of California, Irvine Stanford University
Stanford University University of Michigan
University of Michigan Cornell University
Cornell University University of California, San Diego
University of California, San Diego McGill University
McGill University Northwestern UniversityUniversity of Missouri
Northwestern UniversityUniversity of Missouri University of PennsylvaniaMassachusetts General HospitalUppsala University
University of PennsylvaniaMassachusetts General HospitalUppsala University Duke UniversityHelmholtz MunichMayo ClinicWeill Cornell MedicineIran University of Medical SciencesChildren’s National HospitalUniversity Hospital UlmNational Institute of HealthSage BionetworksUniversity of Rochester Medical CenterMercy Catholic Medical CenterKlinikum rechts der Isar, Technical University of MunichThe Children’s Hospital of PhiladelphiaC.M.H. Lahore Medical College & Institute of DentistryRoss University School of MedicineNassau Medical CenterUniversity of LavalIndiana UniveristyHospital Garcia de Orta - Unidade Local de Sa ́ude de Almada Seixal, EPEScripps Clinic Medical GroupHospital da Luz LisboaVA San Diego Medical CenterASST Ovest Milanese - Legnano
Duke UniversityHelmholtz MunichMayo ClinicWeill Cornell MedicineIran University of Medical SciencesChildren’s National HospitalUniversity Hospital UlmNational Institute of HealthSage BionetworksUniversity of Rochester Medical CenterMercy Catholic Medical CenterKlinikum rechts der Isar, Technical University of MunichThe Children’s Hospital of PhiladelphiaC.M.H. Lahore Medical College & Institute of DentistryRoss University School of MedicineNassau Medical CenterUniversity of LavalIndiana UniveristyHospital Garcia de Orta - Unidade Local de Sa ́ude de Almada Seixal, EPEScripps Clinic Medical GroupHospital da Luz LisboaVA San Diego Medical CenterASST Ovest Milanese - LegnanoGliomas are the most common malignant primary brain tumors in adults and one of the deadliest types of cancer. There are many challenges in treatment and monitoring due to the genetic diversity and high intrinsic heterogeneity in appearance, shape, histology, and treatment response. Treatments include surgery, radiation, and systemic therapies, with magnetic resonance imaging (MRI) playing a key role in treatment planning and post-treatment longitudinal assessment. The 2024 Brain Tumor Segmentation (BraTS) challenge on post-treatment glioma MRI will provide a community standard and benchmark for state-of-the-art automated segmentation models based on the largest expert-annotated post-treatment glioma MRI dataset. Challenge competitors will develop automated segmentation models to predict four distinct tumor sub-regions consisting of enhancing tissue (ET), surrounding non-enhancing T2/fluid-attenuated inversion recovery (FLAIR) hyperintensity (SNFH), non-enhancing tumor core (NETC), and resection cavity (RC). Models will be evaluated on separate validation and test datasets using standardized performance metrics utilized across the BraTS 2024 cluster of challenges, including lesion-wise Dice Similarity Coefficient and Hausdorff Distance. Models developed during this challenge will advance the field of automated MRI segmentation and contribute to their integration into clinical practice, ultimately enhancing patient care.
Researchers from Apple, Cornell, and Weill Cornell Medicine developed PRISM-Bench, a new benchmark using puzzle-based visual tasks to diagnose the step-by-step reasoning fidelity of Multimodal Large Language Models (MLLMs). It revealed that state-of-the-art MLLMs achieve around 50-60% accuracy in identifying a single logical error in a multi-step solution, and that general puzzle-solving ability is only moderately correlated with error detection.
Effectively applying Vision-Language Models (VLMs) to Video Question Answering (VideoQA) hinges on selecting a concise yet comprehensive set of frames, as processing entire videos is computationally infeasible. However, current frame selection methods face a critical trade-off: approaches relying on lightweight similarity models, such as CLIP, often fail to capture the nuances of complex queries, resulting in inaccurate similarity scores that cannot reflect the authentic query-frame relevance, which further undermines frame selection. Meanwhile, methods that leverage a VLM for deeper analysis achieve higher accuracy but incur prohibitive computational costs. To address these limitations, we propose A.I.R., a training-free approach for Adaptive, Iterative, and Reasoning-based frame selection. We leverage a powerful VLM to perform deep, semantic analysis on complex queries, and this analysis is deployed within a cost-effective iterative loop that processes only a small batch of the most high-potential frames at a time. Extensive experiments on various VideoQA benchmarks demonstrate that our approach outperforms existing frame selection methods, significantly boosts the performance of the foundation VLM, and achieves substantial gains in computational efficiency over other VLM-based techniques.
 ETH Zurich
ETH Zurich UCLA
UCLA New York UniversityWeill Cornell MedicineIISER BerhampurIndian Institute of Technology HyderabadCedars-Sinai Medical CenterOakland UniversityUzhhorod National UniversityOregon Health and Science UniversitySage BionetworksTechnical University of Moldova"Stefan cel Mare" University of SuceavaELIXIR Hub
New York UniversityWeill Cornell MedicineIISER BerhampurIndian Institute of Technology HyderabadCedars-Sinai Medical CenterOakland UniversityUzhhorod National UniversityOregon Health and Science UniversitySage BionetworksTechnical University of Moldova"Stefan cel Mare" University of SuceavaELIXIR HubContinuous and reliable access to curated biological data repositories is indispensable for accelerating rigorous scientific inquiry and fostering reproducible research. Centralized repositories, though widely used, are vulnerable to single points of failure arising from cyberattacks, technical faults, natural disasters, or funding and political uncertainties. This can lead to widespread data unavailability, data loss, integrity compromises, and substantial delays in critical research, ultimately impeding scientific progress. Centralizing essential scientific resources in a single geopolitical or institutional hub is inherently dangerous, as any disruption can paralyze diverse ongoing research. The rapid acceleration of data generation, combined with an increasingly volatile global landscape, necessitates a critical re-evaluation of the sustainability of centralized models. Implementing federated and decentralized architectures presents a compelling and future-oriented pathway to substantially strengthen the resilience of scientific data infrastructures, thereby mitigating vulnerabilities and ensuring the long-term integrity of data. Here, we examine the structural limitations of centralized repositories, evaluate federated and decentralized models, and propose a hybrid framework for resilient, FAIR, and sustainable scientific data stewardship. Such an approach offers a significant reduction in exposure to governance instability, infrastructural fragility, and funding volatility, and also fosters fairness and global accessibility. The future of open science depends on integrating these complementary approaches to establish a globally distributed, economically sustainable, and institutionally robust infrastructure that safeguards scientific data as a public good, further ensuring continued accessibility, interoperability, and preservation for generations to come.
04 Sep 2025
The explosion of scientific literature has made the efficient and accurate extraction of structured data a critical component for advancing scientific knowledge and supporting evidence-based decision-making. However, existing tools often struggle to extract and structure multimodal, varied, and inconsistent information across documents into standardized formats. We introduce SciDaSynth, a novel interactive system powered by large language models (LLMs) that automatically generates structured data tables according to users' queries by integrating information from diverse sources, including text, tables, and figures. Furthermore, SciDaSynth supports efficient table data validation and refinement, featuring multi-faceted visual summaries and semantic grouping capabilities to resolve cross-document data inconsistencies. A within-subjects study with nutrition and NLP researchers demonstrates SciDaSynth's effectiveness in producing high-quality structured data more efficiently than baseline methods. We discuss design implications for human-AI collaborative systems supporting data extraction tasks. The system code is available at this https URL
 University of Washington
University of Washington University of Illinois at Urbana-Champaign
University of Illinois at Urbana-Champaign New York UniversityHarvard Medical SchoolMassachusetts General HospitalWeill Cornell MedicineUT Southwestern Medical CenterUniversity of Pittsburgh Medical CenterNational Institutes of HealthNational Library of MedicineOSF HealthCare Illinois Neurological InstituteNational Eye InstituteChungbuk National University College of MedicineChungbuk National University HospitalNorthwestern University, Feinberg School of Medicine
New York UniversityHarvard Medical SchoolMassachusetts General HospitalWeill Cornell MedicineUT Southwestern Medical CenterUniversity of Pittsburgh Medical CenterNational Institutes of HealthNational Library of MedicineOSF HealthCare Illinois Neurological InstituteNational Eye InstituteChungbuk National University College of MedicineChungbuk National University HospitalNorthwestern University, Feinberg School of MedicineSystematic literature review is essential for evidence-based medicine, requiring comprehensive analysis of clinical trial publications. However, the application of artificial intelligence (AI) models for medical literature mining has been limited by insufficient training and evaluation across broad therapeutic areas and diverse tasks. Here, we present LEADS, an AI foundation model for study search, screening, and data extraction from medical literature. The model is trained on 633,759 instruction data points in LEADSInstruct, curated from 21,335 systematic reviews, 453,625 clinical trial publications, and 27,015 clinical trial registries. We showed that LEADS demonstrates consistent improvements over four cutting-edge generic large language models (LLMs) on six tasks. Furthermore, LEADS enhances expert workflows by providing supportive references following expert requests, streamlining processes while maintaining high-quality results. A study with 16 clinicians and medical researchers from 14 different institutions revealed that experts collaborating with LEADS achieved a recall of 0.81 compared to 0.77 experts working alone in study selection, with a time savings of 22.6%. In data extraction tasks, experts using LEADS achieved an accuracy of 0.85 versus 0.80 without using LEADS, alongside a 26.9% time savings. These findings highlight the potential of specialized medical literature foundation models to outperform generic models, delivering significant quality and efficiency benefits when integrated into expert workflows for medical literature mining.
Recent advances in multi-modal AI have demonstrated promising potential for generating the currently expensive spatial transcriptomics (ST) data directly from routine histology images, offering a means to reduce the high cost and time-intensive nature of ST data acquisition. However, the increasing resolution of ST, particularly with platforms such as Visium HD achieving 8um or finer, introduces significant computational and modeling challenges. Conventional spot-by-spot sequential regression frameworks become inefficient and unstable at this scale, while the inherent extreme sparsity and low expression levels of high-resolution ST further complicate both prediction and evaluation. To address these limitations, we propose Img2ST-Net, a novel histology-to-ST generation framework for efficient and parallel high-resolution ST prediction. Unlike conventional spot-by-spot inference methods, Img2ST-Net employs a fully convolutional architecture to generate dense, HD gene expression maps in a parallelized manner. By modeling HD ST data as super-pixel representations, the task is reformulated from image-to-omics inference into a super-content image generation problem with hundreds or thousands of output channels. This design not only improves computational efficiency but also better preserves the spatial organization intrinsic to spatial omics data. To enhance robustness under sparse expression patterns, we further introduce SSIM-ST, a structural-similarity-based evaluation metric tailored for high-resolution ST analysis. We present a scalable, biologically coherent framework for high-resolution ST prediction. Img2ST-Net offers a principled solution for efficient and accurate ST inference at scale. Our contributions lay the groundwork for next-generation ST modeling that is robust and resolution-aware. The source code has been made publicly available at this https URL.
ClinicalBench, a new comprehensive benchmark, systematically evaluates large language models against traditional machine learning models for common clinical prediction tasks. The study demonstrates that large language models, whether directly prompted, engineered, or fine-tuned, currently do not match the predictive performance of traditional models in real-world clinical scenarios.
AGENTiGraph introduces an interactive knowledge graph platform that enables natural language interaction with complex data structures, making knowledge graphs accessible to users without technical expertise. The platform employs a multi-agent architecture to process natural language queries, transforming them into structured graph operations and providing grounded, factual responses across various domains.
We propose a unified framework for adaptive routing in multitask, multimodal prediction settings where data heterogeneity and task interactions vary across samples. Motivated by applications in psychotherapy where structured assessments and unstructured clinician notes coexist with partially missing data and correlated outcomes, we introduce a routing-based architecture that dynamically selects modality processing pathways and task-sharing strategies on a per-sample basis. Our model defines multiple modality paths, including raw and fused representations of text and numeric features and learns to route each input through the most informative expert combination. Task-specific predictions are produced by shared or independent heads depending on the routing decision, and the entire system is trained end-to-end. We evaluate the model on both synthetic data and real-world psychotherapy notes predicting depression and anxiety outcomes. Our experiments show that our method consistently outperforms fixed multitask or single-task baselines, and that the learned routing policy provides interpretable insights into modality relevance and task structure. This addresses critical challenges in personalized healthcare by enabling per-subject adaptive information processing that accounts for data heterogeneity and task correlations. Applied to psychotherapy, this framework could improve mental health outcomes, enhance treatment assignment precision, and increase clinical cost-effectiveness through personalized intervention strategies.
25 Jul 2025
A framework for 'human physiologically-based drug discovery' introduces Programmable Virtual Humans (PVH), which are multiscale computational models designed to simulate drug actions from molecular to phenotypic levels within a virtual human body. This approach aims to directly optimize drug candidates for therapeutic efficacy and minimize side effects *in silico*, addressing the translational gap inherent in traditional drug development.
Breast cancer is one of the leading causes of mortality among women
worldwide. Early detection and risk assessment play a crucial role in improving
survival rates. Therefore, annual or biennial mammograms are often recommended
for screening in high-risk groups. Mammograms are typically interpreted by
expert radiologists based on the Breast Imaging Reporting and Data System
(BI-RADS), which provides a uniform way to describe findings and categorizes
them to indicate the level of concern for breast cancer. Recently, machine
learning (ML) and computational approaches have been developed to automate and
improve the interpretation of mammograms. However, both BI-RADS and the
ML-based methods focus on the analysis of data from the present and sometimes
the most recent prior visit. While it is clear that temporal changes in image
features of the longitudinal scans should carry value for quantifying breast
cancer risk, no prior work has conducted a systematic study of this. In this
paper, we extend a state-of-the-art ML model to ingest an arbitrary number of
longitudinal mammograms and predict future breast cancer risk. On a large-scale
dataset, we demonstrate that our model, LoMaR, achieves state-of-the-art
performance when presented with only the present mammogram. Furthermore, we use
LoMaR to characterize the predictive value of prior visits. Our results show
that longer histories (e.g., up to four prior annual mammograms) can
significantly boost the accuracy of predicting future breast cancer risk,
particularly beyond the short-term. Our code and model weights are available at
this https URL
Fine-tuning Vision Language Models with Graph-based Knowledge for Explainable Medical Image Analysis
Fine-tuning Vision Language Models with Graph-based Knowledge for Explainable Medical Image Analysis
Accurate staging of Diabetic Retinopathy (DR) is essential for guiding timely interventions and preventing vision loss. However, current staging models are hardly interpretable, and most public datasets contain no clinical reasoning or interpretation beyond image-level labels. In this paper, we present a novel method that integrates graph representation learning with vision-language models (VLMs) to deliver explainable DR diagnosis. Our approach leverages optical coherence tomography angiography (OCTA) images by constructing biologically informed graphs that encode key retinal vascular features such as vessel morphology and spatial connectivity. A graph neural network (GNN) then performs DR staging while integrated gradients highlight critical nodes and edges and their individual features that drive the classification decisions. We collect this graph-based knowledge which attributes the model's prediction to physiological structures and their characteristics. We then transform it into textual descriptions for VLMs. We perform instruction-tuning with these textual descriptions and the corresponding image to train a student VLM. This final agent can classify the disease and explain its decision in a human interpretable way solely based on a single image input. Experimental evaluations on both proprietary and public datasets demonstrate that our method not only improves classification accuracy but also offers more clinically interpretable results. An expert study further demonstrates that our method provides more accurate diagnostic explanations and paves the way for precise localization of pathologies in OCTA images.
We present a keypoint-based foundation model for general purpose brain MRI registration, based on the recently-proposed KeyMorph framework. Our model, called BrainMorph, serves as a tool that supports multi-modal, pairwise, and scalable groupwise registration. BrainMorph is trained on a massive dataset of over 100,000 3D volumes, skull-stripped and non-skull-stripped, from nearly 16,000 unique healthy and diseased subjects. BrainMorph is robust to large misalignments, interpretable via interrogating automatically-extracted keypoints, and enables rapid and controllable generation of many plausible transformations with different alignment types and different degrees of nonlinearity at test-time. We demonstrate the superiority of BrainMorph in solving 3D rigid, affine, and nonlinear registration on a variety of multi-modal brain MRI scans of healthy and diseased subjects, in both the pairwise and groupwise setting. In particular, we show registration accuracy and speeds that surpass many classical and learning-based methods, especially in the context of large initial misalignments and large group settings. All code and models are available at this https URL.
With generative artificial intelligence (AI), particularly large language models (LLMs), continuing to make inroads in healthcare, it is critical to supplement traditional automated evaluations with human evaluations. Understanding and evaluating the output of LLMs is essential to assuring safety, reliability, and effectiveness. However, human evaluation's cumbersome, time-consuming, and non-standardized nature presents significant obstacles to comprehensive evaluation and widespread adoption of LLMs in practice. This study reviews existing literature on human evaluation methodologies for LLMs in healthcare. We highlight a notable need for a standardized and consistent human evaluation approach. Our extensive literature search, adhering to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines, includes publications from January 2018 to February 2024. The review examines the human evaluation of LLMs across various medical specialties, addressing factors such as evaluation dimensions, sample types and sizes, selection, and recruitment of evaluators, frameworks and metrics, evaluation process, and statistical analysis type. Drawing on the diverse evaluation strategies employed in these studies, we propose a comprehensive and practical framework for human evaluation of LLMs: QUEST: Quality of Information, Understanding and Reasoning, Expression Style and Persona, Safety and Harm, and Trust and Confidence. This framework aims to improve the reliability, generalizability, and applicability of human evaluation of LLMs in different healthcare applications by defining clear evaluation dimensions and offering detailed guidelines.
The paper provides the first comprehensive survey on Graph Mamba, a novel paradigm integrating State-Space Models (SSMs) with graph learning to address limitations of traditional Graph Neural Networks (GNNs). This integration yields models with linear time complexity and efficient long-range dependency modeling, demonstrating competitive or superior performance across diverse applications, including traffic forecasting, healthcare, and financial markets.
02 Feb 2025
This paper offers a comprehensive review of Healthcare Knowledge Graphs (HKGs), detailing their construction methods, utilization models, and diverse applications across biomedical research, pharmaceutical development, clinical decision support, and public health. It synthesizes existing literature, highlights the integration with Large Language Models (LLMs), and outlines future research directions.
Learning against label noise is a vital topic to guarantee a reliable
performance for deep neural networks. Recent research usually refers to dynamic
noise modeling with model output probabilities and loss values, and then
separates clean and noisy samples. These methods have gained notable success.
However, unlike cherry-picked data, existing approaches often cannot perform
well when facing imbalanced datasets, a common scenario in the real world. We
thoroughly investigate this phenomenon and point out two major issues that
hinder the performance, i.e., \emph{inter-class loss distribution discrepancy}
and \emph{misleading predictions due to uncertainty}. The first issue is that
existing methods often perform class-agnostic noise modeling. However, loss
distributions show a significant discrepancy among classes under class
imbalance, and class-agnostic noise modeling can easily get confused with noisy
samples and samples in minority classes. The second issue refers to that models
may output misleading predictions due to epistemic uncertainty and aleatoric
uncertainty, thus existing methods that rely solely on the output probabilities
may fail to distinguish confident samples. Inspired by our observations, we
propose an Uncertainty-aware Label Correction framework~(ULC) to handle label
noise on imbalanced datasets. First, we perform epistemic uncertainty-aware
class-specific noise modeling to identify trustworthy clean samples and
refine/discard highly confident true/corrupted labels. Then, we introduce
aleatoric uncertainty in the subsequent learning process to prevent noise
accumulation in the label noise modeling process. We conduct experiments on
several synthetic and real-world datasets. The results demonstrate the
effectiveness of the proposed method, especially on imbalanced datasets.
By mapping sites at large scales using remotely sensed data, archaeologists can generate unique insights into long-term demographic trends, inter-regional social networks, and past adaptations to climate change. Remote sensing surveys complement field-based approaches, and their reach can be especially great when combined with deep learning and computer vision techniques. However, conventional supervised deep learning methods face challenges in annotating fine-grained archaeological features at scale. While recent vision foundation models have shown remarkable success in learning large-scale remote sensing data with minimal annotations, most off-the-shelf solutions are designed for RGB images rather than multi-spectral satellite imagery, such as the 8-band data used in our study. In this paper, we introduce DeepAndes, a transformer-based vision foundation model trained on three million multi-spectral satellite images, specifically tailored for Andean archaeology. DeepAndes incorporates a customized DINOv2 self-supervised learning algorithm optimized for 8-band multi-spectral imagery, marking the first foundation model designed explicitly for the Andes region. We evaluate its image understanding performance through imbalanced image classification, image instance retrieval, and pixel-level semantic segmentation tasks. Our experiments show that DeepAndes achieves superior F1 scores, mean average precision, and Dice scores in few-shot learning scenarios, significantly outperforming models trained from scratch or pre-trained on smaller datasets. This underscores the effectiveness of large-scale self-supervised pre-training in archaeological remote sensing. Codes will be available on this https URL.
There are no more papers matching your filters at the moment.